fast BAM/CRAM depth calculation for WGS, exome, or targeted sequencing.
mosdepth can output:
- per-base depth about 2x as fast
samtools depth--about 25 minutes of CPU time for a 30X genome. - mean per-window depth given a window size--as would be used for CNV calling.
- the mean per-region given a BED file of regions.
- the mean or median per-region cumulative coverage histogram given a window size
- a distribution of proportion of bases covered at or above a given threshold for each chromosome and genome-wide.
- quantized output that merges adjacent bases as long as they fall in the same coverage bins e.g. (10-20)
- threshold output to indicate how many bases in each region are covered at the given thresholds.
- A summary of mean depths per chromosome and within specified regions per chromosome.
- a d4 file (better than bigwig).
when appropriate, the output files are bgzipped and indexed for ease of use.
mosdepth 0.3.6
Usage: mosdepth [options] <prefix> <BAM-or-CRAM>
Arguments:
<prefix> outputs: `{prefix}.mosdepth.global.dist.txt`
`{prefix}.mosdepth.summary.txt`
`{prefix}.per-base.bed.gz` (unless -n/--no-per-base is specified)
`{prefix}.regions.bed.gz` (if --by is specified)
`{prefix}.quantized.bed.gz` (if --quantize is specified)
`{prefix}.thresholds.bed.gz` (if --thresholds is specified)
<BAM-or-CRAM> the alignment file for which to calculate depth.
Common Options:
-t --threads <threads> number of BAM decompression threads [default: 0]
-c --chrom <chrom> chromosome to restrict depth calculation.
-b --by <bed|window> optional BED file or (integer) window-sizes.
-n --no-per-base dont output per-base depth. skipping this output will speed execution
substantially. prefer quantized or thresholded values if possible.
-f --fasta <fasta> fasta file for use with CRAM files [default: ].
--d4 output per-base depth in d4 format.
Other options:
-F --flag <FLAG> exclude reads with any of the bits in FLAG set [default: 1796]
-i --include-flag <FLAG> only include reads with any of the bits in FLAG set. default is unset. [default: 0]
-x --fast-mode dont look at internal cigar operations or correct mate overlaps (recommended for most use-cases).
-q --quantize <segments> write quantized output see docs for description.
-Q --mapq <mapq> mapping quality threshold. reads with a quality less than this value are ignored [default: 0]
-l --min-frag-len <min-frag-len> minimum insert size. reads with a smaller insert size than this are ignored [default: -1]
-u --max-frag-len <max-frag-len> maximum insert size. reads with a larger insert size than this are ignored. [default: -1]
-T --thresholds <thresholds> for each interval in --by, write number of bases covered by at
least threshold bases. Specify multiple integer values separated
by ','.
-m --use-median output median of each region (in --by) instead of mean.
-R --read-groups <string> only calculate depth for these comma-separated read groups IDs.
-h --help show help
If you use mosdepth please cite the publication in bioinformatics
See the section below for more info on distribution.
If --by is a BED file with 4 or more columns, it is assumed the the 4th column is the name.
That name will be propagated to the mosdepth output in the 4th column with the depth in the 5th column.
If you don't want this behavior, simply send a bed file with 3 columns.
To calculate the coverage in each exome capture region:
mosdepth --by capture.bed sample-output sample.exome.bam
For a 5.5GB exome BAM and all 1,195,764 ensembl exons as the regions, this completes in 1 minute 38 seconds with a single CPU.
Per-base output will go to sample-output.per-base.bed.gz,
the mean for each region will go to sample-output.regions.bed.gz;
each of those will be written along with a CSI index that can be
used for tabix queries.
The distribution of depths will go to sample-output.mosdepth.dist.txt
For 500-base windows
mosdepth -n --fast-mode --by 500 sample.wgs $sample.wgs.cram
-n means don't output per-base data, this will make mosdepth
a bit faster as there is some cost to outputting that much text.
--fast-mode avoids the extra calculations of mate pair overlap and cigar operations, and also allows htslib to extract less data from CRAM, providing a substantial speed improvement.
To create a set of "callable" regions as in GATK's callable loci tool:
# by setting these ENV vars, we can control the output labels (4th column)
export MOSDEPTH_Q0=NO_COVERAGE # 0 -- defined by the arguments to --quantize
export MOSDEPTH_Q1=LOW_COVERAGE # 1..4
export MOSDEPTH_Q2=CALLABLE # 5..149
export MOSDEPTH_Q3=HIGH_COVERAGE # 150 ...
mosdepth -n --quantize 0:1:5:150: $sample.quantized $sample.wgs.bam
For this case. A regions with depth of 0 are labelled as "NO_COVERAGE", those with coverage of 1,2,3,4 are labelled as "LOW_COVERAGE" and so on.
The result is a BED file where adjacent bases with depths that fall into the same bin are merged into a single region with the 4th column indicating the label.
To get only the distribution value, without the depth file or the per-base and using 3 threads:
MOSDEPTH_PRECISION=5 mosdepth -n -t 3 $sample $bam
Output will go to $sample.mosdepth.dist.txt
This also forces the output to have 5 decimals of precision rather than the default of 2.
D4 is a format created by Hao Hou in the Quinlan lab. It is
incorporated into mosdepth as of version 0.3.0 for per-base output with the --d4 flag.
It improves write speed dramatically; for one test-case it takes 24.8s to write a
per-base.bed.gz with mosdepth compared to 7.7s to write a d4 file. For the same case,
running mosdepth without writing per-base takes 5.9 seconds so D4 greatly mitigates
the cost of outputing per-base depth and the output is more useful.
The simplest option is to download the binary from the releases.
It can also be installed with brew as brew install brewsci/bio/mosdepth or used via docker with quay:
docker pull quay.io/biocontainers/mosdepth:0.3.3--h37c5b7d_2
docker run -v /hostpath/:/opt/mount quay.io/biocontainers/mosdepth:0.2.4--he527e40_0 mosdepth -n --fast-mode -t 4 --by 1000 /opt/mount/sample /opt/mount/$bam
The binary from releases is static, with no dependencies. If you build it yourself,
mosdepth requires htslib version 1.4 or later. If you get an error
about "libhts.so not found", set LD_LIBRARY_PATH to the directory that
contains libhts.so. e.g.
LD_LIBRARY_PATH=~/src/htslib/ mosdepth -h
If you get the error could not import: hts_check_EOF you may need to
install a more recent version of htslib.
If you do want to install from source, see the install.sh.
If you use archlinux, you can install as a package
This is useful for QC.
The $prefix.mosdepth.global.dist.txt file contains, a cumulative distribution indicating the
proportion of total bases (or the proportion of the --by for $prefix.mosdepth.region.dist.txt) that were covered
for at least a given coverage value. It does this for each chromosome, and for the
whole genome.
Each row will indicate:
- chromosome (or "total")
- coverage level
- proportion of bases covered at that level
The last value in each chromosome will be coverage level of 0 aligned with 1.0 bases covered at that level.
A python plotting script is provided in scripts/plot-dist.py that will make
plots like below. Use is python scripts/plot-dist.py \*global.dist.txt and the output
is dist.html with a plot for the full set along with one for each chromosome.
Using something like that, we can plot the distribution from the entire genome. Below we show this for samples with ~60X coverage:
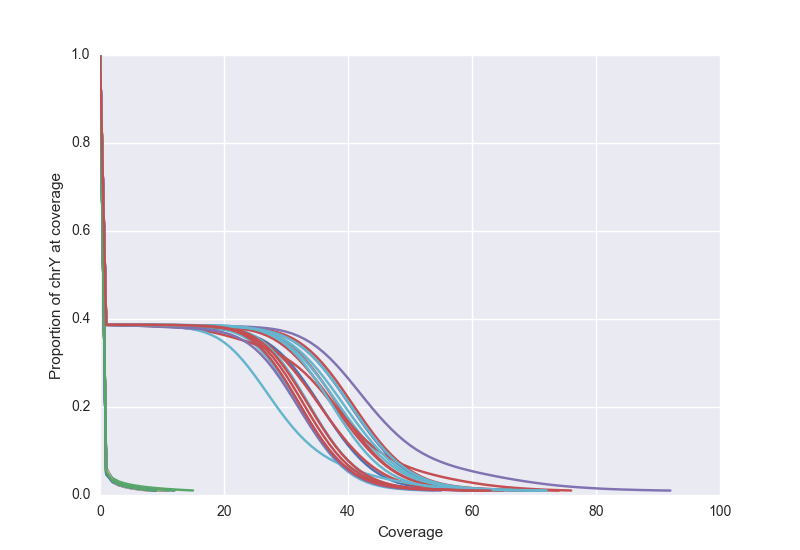
We can also view the Y chromosome to verify that males and females track separately. Below, we that see female samples cluster along the axes while male samples have close to 30X coverage for almost 40% of the genome.
See this blog post for more details.
given a set of regions to the --by argment, mosdepth can report the number of bases in each region that
are covered at or above each threshold value given to --thresholds. e.g:
mosdepth --by exons.bed --thresholds 1,10,20,30 $prefix $bam
will create a file $prefix.thresholds.bed.gz with an extra column for each requested threshold. An example output for the above command (assuming exons.bed had a 4th column with gene names) would look like (including the header):
#chrom start end region 1X 10X 20X 30X
1 11869 12227 ENSE00002234944 358 157 110 0
1 11874 12227 ENSE00002269724 353 127 10 0
1 12010 12057 ENSE00001948541 47 8 0 0
1 12613 12721 ENSE00003582793 108 0 0 0
If there is no name (4th) column in the bed file send to --by then that column will contain "unknown"
in the output.
This is extremely efficient. In our tests, excluding per-base output (-n) and using this argument with
111K exons and 12 values to --thresholds increases the run-time by < 5%.
quantize allows splitting coverage into bins and merging adjacent regions that fall into the same bin even if they have different exact coverage values. This can dramatically reduce the size of the output compared to the per-base.
It also allows outputting regions of low, high, and "callable" coverage as in GATK's callable loci tool.
An example of quantize arguments:
--quantize 0:1:4:100:200: # ... arbitary number of quantize bins.
indicates bins of: 0:1, 1:4, 4:100, 100:200, 200:infinity where the upper endpoint is non-inclusive.
The default for mosdepth is to output labels as above (0:1, 1:4, 4:100... etc.)
To change what is reported as the bin number, a user can set environment variables e.g.:
export MOSDEPTH_Q0=NO_COVERAGE
export MOSDEPTH_Q1=LOW_COVERAGE
export MOSDEPTH_Q2=CALLABLE
export MOSDEPTH_Q3=HIGH_COVERAGE
In this case, the bin label is replaced by the text in the appropriate environment variable.
This is very efficient. In our tests, excluding per-base output (-n) and using this argument with
9 bins to --quantize increases the run-time by ~ 20%. In contrast, the difference in time with
and without -n can be 2-fold.
As it encounters each chromosome, mosdepth creates an array the length of the chromosome.
For every start it encounters, it increments the value in that position of the array. For every
stop, it decrements that position. From this, the depth at a particular position is the
cumulative sum of all array positions preceding it (a similar algorithm is used in BEDTools
where starts and stops are tracked separately). mosdepth avoids double-counting
overlapping mate-pairs and it tracks every aligned part of every read using the CIGAR
operations. Because of this data structure, the the coverage distribution calculation
can be done without a noticeable increase in run-time. The image below conveys the concept:
This array accounting is very fast. There are no extra allocations or objects to track and
it is also conceptually simple. For these reasons, it is faster than samtools depth which
works by using the pileup machinery that
tracks each read, each base.
The mosdepth method has some limitations. Because a large array is allocated and it is
required (in general) to take the cumulative sum of all preceding positions to know the depth
at any position, it is slower for small, 1-time regional queries. It is, however fast for
window-based or BED-based regions, because it first calculates the full chromosome coverage
and then reports the coverage for each region in that chromosome. Another downside is it uses
more memory than samtools. The amount of memory is approximately equal to 32-bits * longest chrom
length, so for the 249MB chromosome 1, it will require 1GB of memory.
mosdepth is written in nim and it uses our htslib
via our nim wrapper hts-nim
mosdepth, samtools, bedtools, and sambamba were run on a 30X genome.
relative times are relative to mosdepth per-base mode with a single thread.
mosdepth can report the mean depth in 500-base windows genome-wide info
under 9 minutes of user time with 3 threads.
| format | tool | threads | mode | relative time | run-time | memory |
|---|---|---|---|---|---|---|
| BAM | mosdepth | 1 | base | 1 | 25:23 | 1196 |
| BAM | mosdepth | 3 | base | 0.57 | 14:27 | 1197 |
| CRAM | mosdepth | 1 | base | 1.17 | 29:47 | 1205 |
| CRAM | mosdepth | 3 | base | 0.56 | 14:08 | 1225 |
| BAM | mosdepth | 3 | window | 0.34 | 8:44 | 1277 |
| BAM | mosdepth | 1 | window | 0.80 | 20:26 | 1212 |
| CRAM | mosdepth | 3 | window | 0.35 | 8:47 | 1233 |
| CRAM | mosdepth | 1 | window | 0.88 | 22:23 | 1209 |
| BAM | sambamba | 1 | base | 5.71 | 2:24:53 | 166 |
| BAM | samtools | 1 | base | 1.98 | 50:12 | 27 |
| CRAM | samtools | 1 | base | 1.79 | 45:21 | 451 |
| BAM | bedtools | 1 | base | 5.31 | 2:14:44 | 1908 |
Note that the threads to mosdepth (and samtools) are decompression threads. After
about 4 threads, there is no benefit for additional threads:
We compared samtools depth with default arguments to mosdepth without overlap detection and discovered no
differences across the entire chromosome.