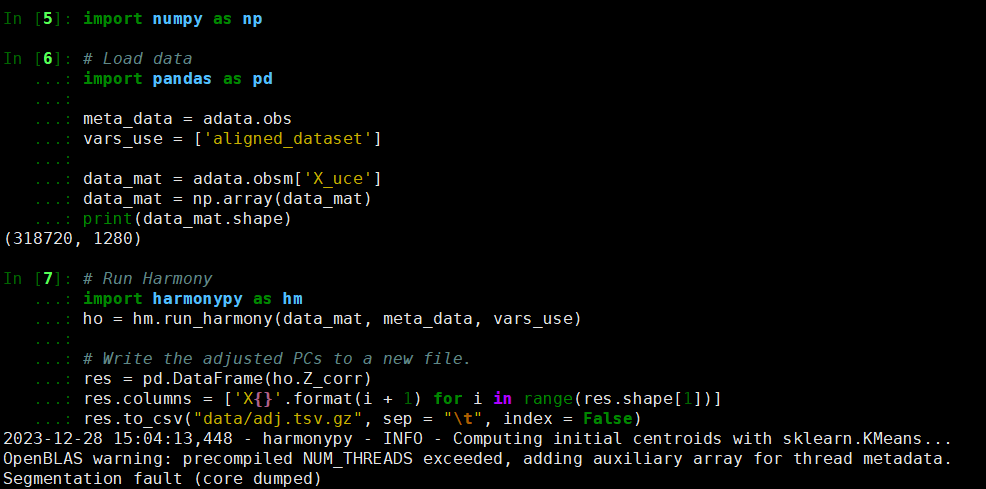
I am trying to run Harmonypy on an anndata object. It gave me the error below.
AttributeError Traceback (most recent call last)
Cell In[198], line 1
----> 1 sc.external.pp.harmony_integrate(xenium_breast_htma_breast, key = 'patient')
File ~/anaconda3/lib/python3.10/site-packages/scanpy/external/pp/_harmony_integrate.py:82, in harmony_integrate(adata, key, basis, adjusted_basis, **kwargs)
79 except ImportError:
80 raise ImportError("\nplease install harmonypy:\n\n\tpip install harmonypy")
---> 82 harmony_out = harmonypy.run_harmony(adata.obsm[basis], adata.obs, key, **kwargs)
84 adata.obsm[adjusted_basis] = harmony_out.Z_corr.T
File ~/anaconda3/lib/python3.10/site-packages/harmonypy/harmony.py:124, in run_harmony(data_mat, meta_data, vars_use, theta, lamb, sigma, nclust, tau, block_size, max_iter_harmony, max_iter_kmeans, epsilon_cluster, epsilon_harmony, plot_convergence, verbose, reference_values, cluster_prior, random_state)
120 phi_moe = np.vstack((np.repeat(1, N), phi))
122 np.random.seed(random_state)
--> 124 ho = Harmony(
125 data_mat, phi, phi_moe, Pr_b, sigma, theta, max_iter_harmony, max_iter_kmeans,
126 epsilon_cluster, epsilon_harmony, nclust, block_size, lamb_mat, verbose
127 )
129 return ho
File ~/anaconda3/lib/python3.10/site-packages/harmonypy/harmony.py:172, in Harmony.__init__(self, Z, Phi, Phi_moe, Pr_b, sigma, theta, max_iter_harmony, max_iter_kmeans, epsilon_kmeans, epsilon_harmony, K, block_size, lamb, verbose)
169 self.kmeans_rounds = []
171 self.allocate_buffers()
--> 172 self.init_cluster()
173 self.harmonize(self.max_iter_harmony, self.verbose)
File ~/anaconda3/lib/python3.10/site-packages/harmonypy/harmony.py:191, in Harmony.init_cluster(self)
188 logger.info("Computing initial centroids with sklearn.KMeans...")
189 model = KMeans(n_clusters=self.K, init='k-means++',
190 n_init=10, max_iter=25)
--> 191 model.fit(self.Z_cos.T)
192 km_centroids, km_labels = model.cluster_centers_, model.labels_
193 logger.info("sklearn.KMeans initialization complete.")
File ~/anaconda3/lib/python3.10/site-packages/sklearn/cluster/_kmeans.py:1468, in KMeans.fit(self, X, y, sample_weight)
1465 print("Initialization complete")
1467 # run a k-means once
-> 1468 labels, inertia, centers, n_iter_ = kmeans_single(
1469 X,
1470 sample_weight,
1471 centers_init,
1472 max_iter=self.max_iter,
1473 verbose=self.verbose,
1474 tol=self._tol,
1475 n_threads=self._n_threads,
1476 )
1478 # determine if these results are the best so far
1479 # we chose a new run if it has a better inertia and the clustering is
1480 # different from the best so far (it's possible that the inertia is
1481 # slightly better even if the clustering is the same with potentially
1482 # permuted labels, due to rounding errors)
1483 if best_inertia is None or (
1484 inertia < best_inertia
1485 and not _is_same_clustering(labels, best_labels, self.n_clusters)
1486 ):
File ~/anaconda3/lib/python3.10/site-packages/sklearn/cluster/_kmeans.py:679, in _kmeans_single_lloyd(X, sample_weight, centers_init, max_iter, verbose, tol, n_threads)
675 strict_convergence = False
677 # Threadpoolctl context to limit the number of threads in second level of
678 # nested parallelism (i.e. BLAS) to avoid oversubscription.
--> 679 with threadpool_limits(limits=1, user_api="blas"):
680 for i in range(max_iter):
681 lloyd_iter(
682 X,
683 sample_weight,
(...)
689 n_threads,
690 )
File ~/anaconda3/lib/python3.10/site-packages/sklearn/utils/fixes.py:139, in threadpool_limits(limits, user_api)
137 return controller.limit(limits=limits, user_api=user_api)
138 else:
--> 139 return threadpoolctl.threadpool_limits(limits=limits, user_api=user_api)
File ~/anaconda3/lib/python3.10/site-packages/threadpoolctl.py:171, in threadpool_limits.__init__(self, limits, user_api)
167 def __init__(self, limits=None, user_api=None):
168 self._limits, self._user_api, self._prefixes = \
169 self._check_params(limits, user_api)
--> 171 self._original_info = self._set_threadpool_limits()
File ~/anaconda3/lib/python3.10/site-packages/threadpoolctl.py:268, in threadpool_limits._set_threadpool_limits(self)
265 if self._limits is None:
266 return None
--> 268 modules = _ThreadpoolInfo(prefixes=self._prefixes,
269 user_api=self._user_api)
270 for module in modules:
271 # self._limits is a dict {key: num_threads} where key is either
272 # a prefix or a user_api. If a module matches both, the limit
273 # corresponding to the prefix is chosed.
274 if module.prefix in self._limits:
File ~/anaconda3/lib/python3.10/site-packages/threadpoolctl.py:340, in _ThreadpoolInfo.__init__(self, user_api, prefixes, modules)
337 self.user_api = [] if user_api is None else user_api
339 self.modules = []
--> 340 self._load_modules()
341 self._warn_if_incompatible_openmp()
342 else:
File ~/anaconda3/lib/python3.10/site-packages/threadpoolctl.py:371, in _ThreadpoolInfo._load_modules(self)
369 """Loop through loaded libraries and store supported ones"""
370 if sys.platform == "darwin":
--> 371 self._find_modules_with_dyld()
372 elif sys.platform == "win32":
373 self._find_modules_with_enum_process_module_ex()
File ~/anaconda3/lib/python3.10/site-packages/threadpoolctl.py:428, in _ThreadpoolInfo._find_modules_with_dyld(self)
425 filepath = filepath.decode("utf-8")
427 # Store the module if it is supported and selected
--> 428 self._make_module_from_path(filepath)
File ~/anaconda3/lib/python3.10/site-packages/threadpoolctl.py:515, in _ThreadpoolInfo._make_module_from_path(self, filepath)
513 if prefix in self.prefixes or user_api in self.user_api:
514 module_class = globals()[module_class]
--> 515 module = module_class(filepath, prefix, user_api, internal_api)
516 self.modules.append(module)
File ~/anaconda3/lib/python3.10/site-packages/threadpoolctl.py:606, in _Module.__init__(self, filepath, prefix, user_api, internal_api)
604 self.internal_api = internal_api
605 self._dynlib = ctypes.CDLL(filepath, mode=_RTLD_NOLOAD)
--> 606 self.version = self.get_version()
607 self.num_threads = self.get_num_threads()
608 self._get_extra_info()
File ~/anaconda3/lib/python3.10/site-packages/threadpoolctl.py:646, in _OpenBLASModule.get_version(self)
643 get_config = getattr(self._dynlib, "openblas_get_config",
644 lambda: None)
645 get_config.restype = ctypes.c_char_p
--> 646 config = get_config().split()
647 if config[0] == b"OpenBLAS":
648 return config[1].decode("utf-8")
AttributeError: 'NoneType' object has no attribute 'split'
I tried the method in the closed issue. Didn't work for me. Any help is greatly appreciated!! Thanks.