Original report (archived issue) by Benjamin Brown (Bitbucket: Benjamin Brown).
I've come across an interesting parser/substitution problem with the integ() operator, when applied across the whole domain. This is for a 2-D problem, Fourier ("x") -- Chebyshev ("z").
I defined two averaging types, one over planes perpendicular to the chebyshev direction, and one over the whole domain:
#!python
problem.substitutions['plane_avg(A)'] = 'integ(A, "x")/Lx'
problem.substitutions['vol_avg(A)'] = 'integ(A)/Lx/Lz'
and a substitiution:
#!python
problem.substitutions['enstrophy'] = '(dx(w) - u_z)**2'
After setting up equations, etc. I initialize slice, profile_avg and vol_avg (scalar) outputs, each in their own file:
#!python
analysis_slice = solver.evaluator.add_file_handler(data_dir+"slices", max_writes=20, parallel=False, **kwargs)
analysis_slice.add_task("enstrophy", name="enstrophy")
analysis_profile = solver.evaluator.add_file_handler(data_dir+"profiles", max_writes=20, parallel=False, **kwargs)
analysis_profile.add_task("plane_avg(enstrophy)", name="enstrophy")
analysis_scalar = solver.evaluator.add_file_handler(data_dir+"scalar", max_writes=20, parallel=False, **kwargs)
analysis_scalar.add_task("vol_avg(enstrophy)", name="enstrophy")
That code fails with the following grid-data error:
#!python
Traceback (most recent call last):
File "FC_fixed_kappa_1e6.py", line 111, in <module>
solver.step(dt)
File "/u/bpbrown/build/dedalus/dedalus/core/solvers.py", line 339, in step
self.timestepper.step(self, dt, wall_time)
File "/u/bpbrown/build/dedalus/dedalus/core/timesteppers.py", line 528, in step
evaluator.evaluate_scheduled(wall_time, solver.sim_time, iteration)
File "/u/bpbrown/build/dedalus/dedalus/core/evaluator.py", line 97, in evaluate_scheduled
self.evaluate_handlers(scheduled_handlers, wall_time, sim_time, iteration)
File "/u/bpbrown/build/dedalus/dedalus/core/evaluator.py", line 127, in evaluate_handlers
self.domain.dist.paths[next_index].decrement(fields)
File "/u/bpbrown/build/dedalus/dedalus/core/distributor.py", line 343, in decrement
self.decrement_single(field)
File "/u/bpbrown/build/dedalus/dedalus/core/distributor.py", line 323, in decrement_single
self.basis.forward(gdata, cdata, self.axis, field.meta[self.axis])
File "/u/bpbrown/build/dedalus/dedalus/core/basis.py", line 717, in _forward_fftw
cdata, gdata = self.check_arrays(cdata, gdata, axis)
File "/u/bpbrown/build/dedalus/dedalus/core/basis.py", line 148, in check_arrays
raise ValueError("gdata does not match grid_dtype")
ValueError: gdata does not match grid_dtype
Changing only the very last line of the analysis code block:
#!python
analysis_scalar.add_task("vol_avg(enstrophy)", name="enstrophy")
to do the substitution manually:
#!python
analysis_scalar.add_task("vol_avg((dx(w) - u_z)**2)", name="enstrophy")
causes the code to run fine and trigger no such error.
This seems like some form of substitution/parser bug. Any ideas?






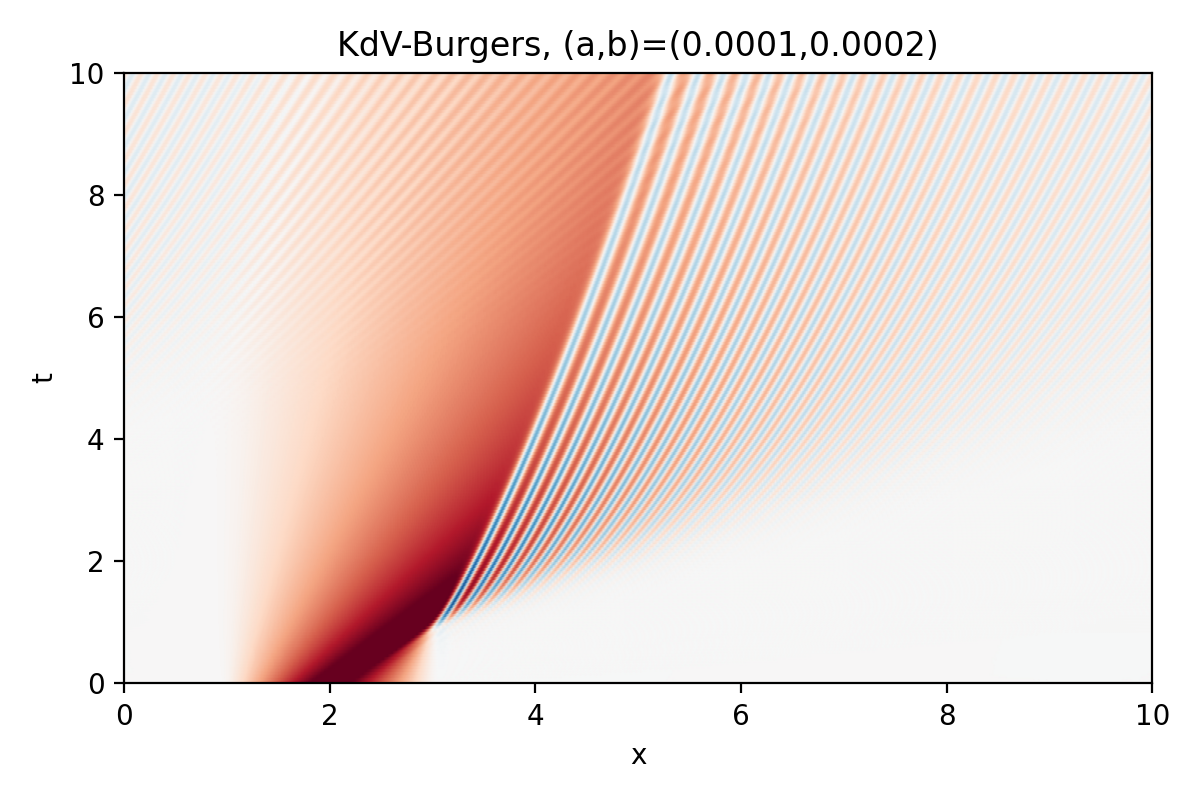 KdV-Burgers equation (1D IVP)
KdV-Burgers equation (1D IVP)
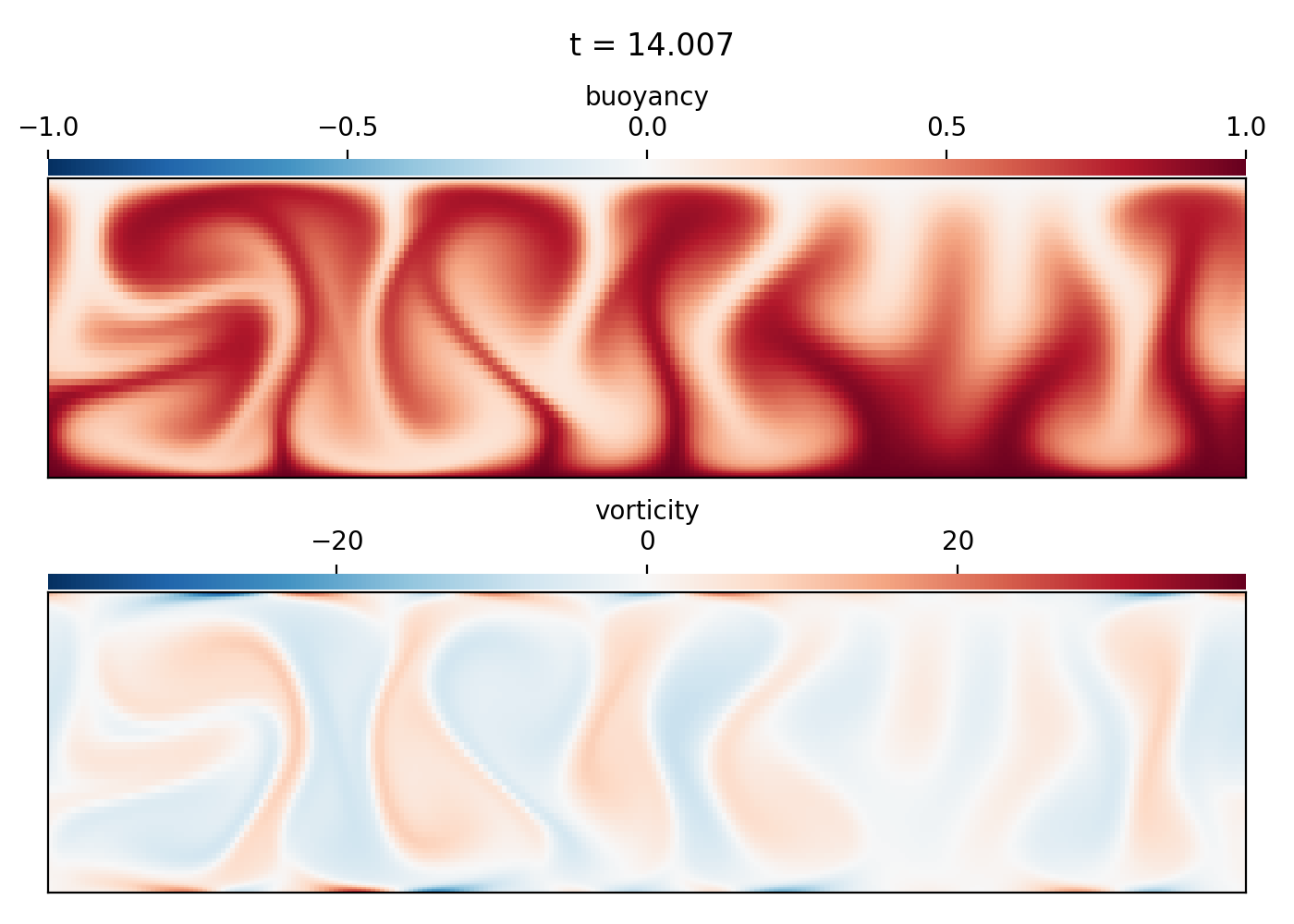 Rayleigh-Benard convection (2D IVP)
Rayleigh-Benard convection (2D IVP)
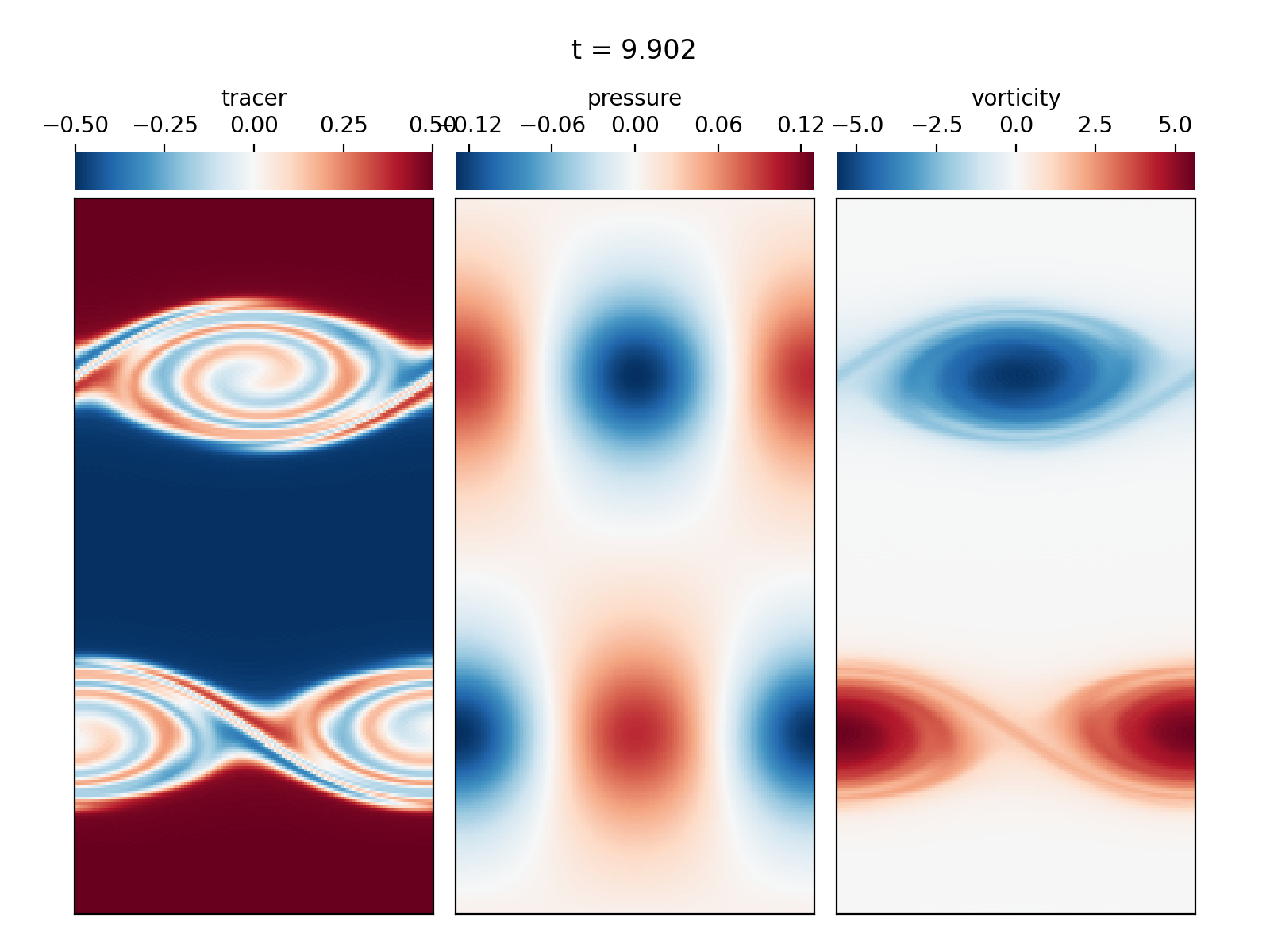 Periodic shear flow (2D IVP)
Periodic shear flow (2D IVP)
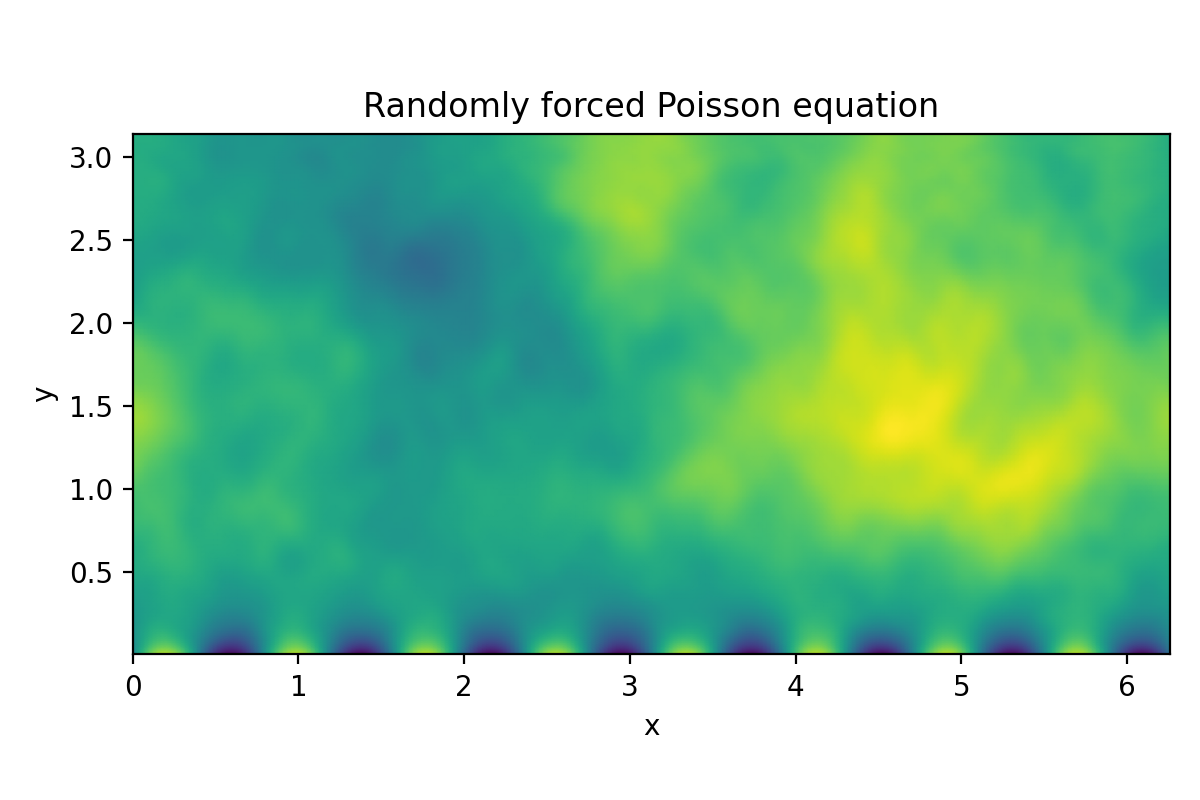 Poisson equation (2D LBVP)
Poisson equation (2D LBVP)
 Librational instability (disk IVP)
Librational instability (disk IVP)
 Spherical shallow water (sphere IVP)
Spherical shallow water (sphere IVP)
 Spherical shell convection (shell IVP)
Spherical shell convection (shell IVP)
 Internally heated convection (ball IVP)
Internally heated convection (ball IVP)