cupy / cupy Goto Github PK
View Code? Open in Web Editor NEWNumPy & SciPy for GPU
Home Page: https://cupy.dev
License: MIT License
NumPy & SciPy for GPU
Home Page: https://cupy.dev
License: MIT License
Some functions implement incompatible arguments with NumPy. We should fix that for more usability.
Fix argument
copy(a, order='k')array(object, dtype=None, copy=True, order='K', subok=False, ndmin=0)Linking from chainer's issue tracker chainer/chainer#3075
In the code below, the seed should be set to 10 no matter what order these threads are executed.
Code:
import threading
import random
import cupy
SEED = 10
def func_seed():
cupy.random.seed(SEED)
def func_get_random_state():
cupy.random.get_random_state()
def test():
procs = [
threading.Thread(target=func_seed),
threading.Thread(target=func_get_random_state),
threading.Thread(target=func_get_random_state),
threading.Thread(target=func_get_random_state),
threading.Thread(target=func_get_random_state),
threading.Thread(target=func_get_random_state),
threading.Thread(target=func_get_random_state),
]
random.shuffle(procs)
for p in procs:
p.start()
#p.join() # this hides the problem
for p in procs:
p.join()
actual = cupy.random.uniform()
cupy.random.seed(SEED)
expected = cupy.random.uniform()
print("Expected: {}".format(expected))
print("Actual : {}".format(actual))
test()
Result:
Expected: 0.6320792449063122
Actual : 0.41662013290707867
Arrays sorted with cupy.sort operation have some properties such as dtype, rank, sorting axis and C/F-contiguousness. Currently, cupy.sort supports sorting arrays only with the rank of one because of its implementation reason, see #55.
This issue addresses a problem that makes cupy.sort support sorting arrays with the rank of two or more, with the last axis and C-contiguousness.
Rank two
For an array with the rank of two,
[[4, 3]
[2, 1]]
treating the array as flattened one, [4, 3, 2 ,1], and providing the following comparator in pseudo code to underlying Thrust library:
if floor(i / 2) < floor(j / 2) then return true;
else if floor(i / 2) > floor(j / 2) then return false;
else return data[i] < data[j];
where i and j are array indices, and data[i] represents i th element of array data,
we get the C-contiguous array sorted with the last axis.
[[3, 4]
[1, 2]]
Rank N
Generalized to the rank of N with shape (d_0, d_1, ..., d_n-1), the following comparator works:
if floor(i / d_n-1) < floor(j / d_n-1) then return true;
else if floor(i / d_n-1) > floor(j / d_n-1) then return false;
else return data[i] < data[j];
cupy.partition, the counterpart of numpy.partition, creates a copy of an array with its elements rearranged in such a way that the value of the element in k-th position is in the position it would be in a sorted array. This function is equivalent to std::nth_element of C++ STL.
numpy.partition
https://docs.scipy.org/doc/numpy/reference/generated/numpy.partition.html
std::nth_element
http://en.cppreference.com/w/cpp/algorithm/nth_element
Unfortunately, neither of the following parallel algorithm libraries implement this kind of parallel selection algorithm:
For now, the most likely is to implement radix select based on CUB's radix sort implementation.
git clone [email protected]:cupy/cupy cupy_test
cd cupy_test
python setup.develop
Modify some pxd files such as:
diff --git a/cupy/cuda/memory.pxd b/cupy/cuda/memory.pxd
index 7c98770..063ed31 100644
--- a/cupy/cuda/memory.pxd
+++ b/cupy/cuda/memory.pxd
@@ -14,6 +14,7 @@ cdef class MemoryPointer:
readonly device.Device device
readonly object mem
readonly size_t ptr
+ readonly int size
cpdef copy_from_device(self, MemoryPointer src, Py_ssize_t size)
cpdef copy_from_device_async(self, MemoryPointer src, size_t size, stream)Then, I get following errors:
$ python setup.py develop
$ python -m unittest tests/cupy_tests/cuda_tests/test_memory.py
...
...
ValueError: cupy.cuda.memory.MemoryPointer has the wrong size, try recompiling. Expected 56, got 48
$ python setup.py clean
$ python setup.py develop
$ python -m unittest tests/cupy_tests/cuda_tests/test_memory.py
...
...
ValueError: cupy.cuda.memory.MemoryPointer has the wrong size, try recompiling. Expected 56, got 48
The problems I have are:
python setup.py develop does not rebuild pxd wellpython setup.py clean does not cleanup everything wellCuPy Overview says:
CuPy also includes following features for performance:
- Customizable memory allocator, and a simple memory pool as an example
When writing a custom memory allocator from scratch, I want to reuse or inherit some classes in CuPy (cupy.cuda.memory.Memory, cupy.cuda.memory.MemoryPointer). However *.pxd files are not installed along with the module by pip install cupy, so cupy modules cannot be cimported from my Cython module. Currently I have to copy *.pxd files from CuPy source tree to my project and specify the path to include_path option when building my module using Cython.
So it would be nice if CuPy installs *.pxd files to make it cimport-able from other packages. I think this is possible by using package_data option as mentioned in the Cython docs:
I post this from #43 (comment).
It would be nice if there were a @fuse(cached=True) or @fuse_cached which enables the kernel generated by one call to the function to be reused for another call.
NCCL is not installed in the current Docker image.
Feature suggestion:
It would be helpful if the content of .cu file is optionally dumped to stderr if there were an NVCC compilation error.
The main usage in my mind is for CI, where you cannot see the source (stored in a temporary file) afterward.
I'm using pip to install this. but there is an error.
Command "/usr/bin/python3 -u -c "import setuptools, tokenize;file='/tmp/pip-build-raeuoelt/cupy/setup.py';f=getattr(tokenize, 'open', open)(file);code=f.read().replace('\r\n', '\n');f.close();exec(compile(code, file, 'exec'))" install --record /tmp/pip-yl91qc3r-record/install-record.txt --single-version-externally-managed --compile" failed with error code 1 in /tmp/pip-build-raeuoelt/cupy/
Traceback
Traceback (most recent call last):
File "/usr/local/lib/python3.5/dist-packages/pip/basecommand.py", line 215, in main
status = self.run(options, args)
File "/usr/local/lib/python3.5/dist-packages/pip/commands/install.py", line 342, in run
prefix=options.prefix_path,
File "/usr/local/lib/python3.5/dist-packages/pip/req/req_set.py", line 784, in install
**kwargs
File "/usr/local/lib/python3.5/dist-packages/pip/req/req_install.py", line 878, in install
spinner=spinner,
File "/usr/local/lib/python3.5/dist-packages/pip/utils/init.py", line 707, in call_subprocess
% (command_desc, proc.returncode, cwd))
eigh and eigvalsh does not support Hermitian matrix because CuPy currently does not support complex though cuSolver support it. We can easily support Hermitian matrix after CuPy support complex.
related to #46
Hi! I have been looking into this package and trying to implement simple image processing filters, like median filter, by writing the kernel for it. This is a test function for looping over the 2D array, it's not fully implemented median filter (the kernel parameter is ignored):
import cupy as cp
def loop2d(data, kernel=3):
"""
This should be doing a row-major traversal.
The data is broadcast into a 1D array when transferred to the device.
"""
# allocate memory for the output parameter
data_out = cp.zeros(data.shape, dtype=data.dtype)
f = cp.ElementwiseKernel(
in_params='raw T in, int32 width, int32 kernel, int32 kernel_width',
out_params='raw T out',
preamble=r"""
#include <stdio.h> // for debug
""",
operation=r"""
printf("index: %d\n", i);
for(int j = 0; j < width; ++j){
int pos = i * width + j;
printf("value at %d: %d\n", pos, in[pos]);
out[pos] = in[pos] + 3; // apply some operation
}
""",
name='loop2d',
options=('-std=c++11',),
reduce_dims=False
)
if kernel <= 1:
return data
kernel_width = (kernel - 1) / 2
# this is for the outer for loop, looping over the rows
data_height = data.shape[1]
# this is for the inner loop, looping over the columns
data_width = data.shape[0]
# we pass in data_out as the output parameter
return f(data, data_width, kernel, kernel_width, data_out, size=data_height)
some_data = cp.full((10,10), 1, dtype=cp.int32)
loop2d(some_data)
I feel this is abusing the ElementwiseKernel, because it's operating row-wise, not element wise.
I am curious to know if my traversal approach is correct? Is there another way to traverse a 2D array, without manually calculating the current position after being broadcast?
How can I find what functions I can use within the CUDA kernel declaration? I have been looking around random files in the source code that use kernels, and functions like atomicAdd that have been used in some of them can only be found within the source files.
Expanding on the previous Is there a way to define our own functions? A hack that works is to define the function within the preamble='void myfunc(){};', but maybe there's a way to add it into cupy in a different way
Thank you very much for your time! I am very impressed with the work that you all have done on this package, so I can only say, keep up the good work!
Fusion code raises an error when out argument is specified.
import cupy
import numpy
xp = cupy
a = xp.ones((2,3))
b = xp.ones((2,3)) * 2
z = xp.zeros((2,3))
@cupy.fuse()
def func(a, b, z):
xp.add(a, b, out=z)
func(a, b, z)
print(z)
Results in:
Traceback (most recent call last):
File "test-out.py", line 13, in <module>
func(a, b, z)
File "/repos/cupy/cupy/core/fusion.py", line 602, in __call__
return self._call(*args, **kwargs)
File "/repos/cupy/cupy/core/fusion.py", line 628, in _call
self.post_map, self.identity, types)
File "/repos/cupy/cupy/core/fusion.py", line 499, in _get_fusion
out_refs = func(*in_refs)
File "test-out.py", line 11, in func
xp.add(a, b, out=z)
File "/repos/cupy/cupy/core/fusion.py", line 710, in __call__
return _convert(self._fusion_op)(*args, **kwargs)
File "/repos/cupy/cupy/core/fusion.py", line 339, in res
var_list.append(_normalize_arg.pop('out'))
AttributeError: 'function' object has no attribute 'pop'
It could be directly fused with ordinary function-call notation.
Although CuPy supports creating CUDA stream, generated kernels always run in the default (null) CUDA stream.
It would be nice if CUDA stream to launch kernels can be specified using context manager, just like cupy.cuda.Device.
The CuPy contribution guide is almost copied from the one from Chainer v1. We should update so that it is consistent with Chainer v2. Currently we are updating the API compatibility policy in Chainer. We will port and modify it to be accommodated to CuPy.
First of all - thank you for this wonderful project!
I use Pandas frequently in my work. I am curious:
This error happens when I try to install CuPy in Ubuntu 17.04
$ python setup.py develop
:
:
NVCC options: ['--generate-code=arch=compute_30,code=compute_30', '--generate-code=arch=compute_50,code=compute_50', '--generate-code=arch=compute_60,code=compute_60', '-O2', '--compiler-options="-fPIC"']
/usr/bin/nvcc -D_GLIBCXX_USE_CXX11_ABI=0 -D_FORCE_INLINES=1 -I/usr/include -I/home/niboshi/anaconda/anaconda3/include/python3.6m -c cupy/cuda/cupy_thrust.cu -o build/temp.linux-x86_64-3.6/cupy/cuda/cupy_thrust.o --generate-code=arch=compute_30,code=compute_30 --generate-code=arch=compute_50,code=compute_50 --generate-code=arch=compute_60,code=compute_60 -O2 --compiler-options="-fPIC"
ERROR: No supported gcc/g++ host compiler found, but clang-3.8 is available.
Use 'nvcc -ccbin clang-3.8' to use that instead.
error: command '/usr/bin/nvcc' failed with exit status 1
I found the behaviour of argmax is different between numpy and cupy if the array has 0 in its shape and axis argument is used. I don't know whether this behaviour is intended or not.
np.empty((0, 1)).argmax(axis=1) # array([], dtype=int64)
cupy.empty((0, 1)).argmax(axis=1)
---------------------------------------------------------------------------
ValueError Traceback (most recent call last)
<ipython-input-9-a5737d72bcba> in <module>()
----> 1 cupy.empty((0, 1)).argmax(axis=1)
cupy/core/core.pyx in cupy.core.core.ndarray.argmax (cupy/core/core.cpp:17701)()
cupy/core/core.pyx in cupy.core.core.ndarray.argmax (cupy/core/core.cpp:17556)()
cupy/core/reduction.pxi in cupy.core.core.simple_reduction_function.__call__ (cupy/core/core.cpp:52697)()
ValueError: zero-size array to reduction operation cupy_argmax which has no identity
I used cupy 2.0.0a1.
When using numpy 1.12.1:
>>> import cupy
>>> cupy.cumprod
<function cumprod at 0x7f9a460b4c80>
>>> cupy.cumprod(cupy.ndarray(()), axis=-10000)
Traceback (most recent call last):
File "<stdin>", line 1, in <module>
File "/path/to/cupy/math/sumprod.py", line 206, in cumprod
raise numpy.AxisError('axis(={}) out of bounds'.format(axis))
AttributeError: module 'numpy' has no attribute 'AxisError'$ echo $PATH
/usr/local/cuda/bin:/home/wkentaro/.local/bin:/usr/local/bin:/usr/local/sbin:/usr/local/sbin:/usr/local/bin:/usr/sbin:/usr/bin:/sbin:/bin:/usr/games:/usr/local/games
$ echo $LD_LIBRARY_PATH
/usr/local/cuda/lib64:
$ echo $CFLAGS
-I/usr/local/cuda/include
$ echo $LDFLAGS
-L/usr/local/cuda/lib64
$ sudo pip install cupy --no-cache-dir -vvv
...
building 'cupy.cuda.thrust' extension
error: unknown file type '.cu' (from 'cupy/cuda/cupy_thrust.cu')
Running setup.py install for cupy: finished with status 'error'
Cleaning up...
It says below unexpectedly.
**************************************************
*** WARNING: nvcc not in path.
*** WARNING: Please set path to nvcc.
**************************************************
Do anyone have some idea to fix this? It worked before I upgrade the cuda to 8.0.61 from 8.0.5X.
Although numpy.nan exists, cupy.nan does not.
Code:
import cupy
@cupy.fuse()
def func(a, b):
pass
x = cupy.ones((2, 2))
y = cupy.ones((2, 2)) *3
z = func(x, y)
print(z)
Results:
[[ 3. 3.]
[ 3. 3.]]
Hi, I'm trying to write a kernel function with ElementwiseKernel.
My code is simple, I put this function in a class:
self.update_params_cuda = cp.ElementwiseKernel(
'float32 m, float32 v,float32 lr, float32 grad',
'float32 u'
'u = m*v-lr*grad',
'update_params_cuda'
)
but when execute to this, no matter float32/ T / raw T raises Unknown keyword error.
Any idea how to fix this?
File "cupy/core/elementwise.pxi", line 466, in cupy.core.core.ElementwiseKernel.__init__ (cupy/core/core.cpp:42249)
File "cupy/util.pyx", line 39, in cupy.util.memoize.decorator.ret (cupy/util.cpp:1481)
File "cupy/core/elementwise.pxi", line 262, in cupy.core.core._get_param_info (cupy/core/core.cpp:38189)
File "cupy/core/elementwise.pxi", line 255, in cupy.core.core.ParameterInfo.__init__ (cupy/core/core.cpp:37751)
Exception: Unknown keyword "float32"
I want to implement sparse matrix named cupy.sparse whose interface is same as scipy.sparse using cuSparse.
I investigated specification of cuSparse.
scipy.sparse.scipy.sparse has no special implementation except for csc_matrix. It convert matrixes to csc format and call its implementation.__array_priority__ attribute to control calling operator method such as __add__.Todo:
spmatrix: base class of all sparse matrixes (https://docs.scipy.org/doc/scipy-0.19.0/reference/generated/scipy.sparse.spmatrix.html#scipy.sparse.spmatrix) (#40)csr_matrix: most basic sparse matrix in cuSparse (https://docs.scipy.org/doc/scipy-0.19.0/reference/generated/scipy.sparse.csr_matrix.html)
csc_matrix: transpose of csr (https://docs.scipy.org/doc/scipy-0.19.0/reference/generated/scipy.sparse.csc_matrix.html#scipy.sparse.csc_matrix) (#226)coo_matrix: it is easier to make (https://docs.scipy.org/doc/scipy-0.19.0/reference/generated/scipy.sparse.coo_matrix.html#scipy.sparse.coo_matrix) (#234)__add__ and __sub__
dot and __mul__
Code:
import sys
import cupy, numpy
shapes = [
(),
(0,),
(1,),
(0,2),
(0,0,2,0),
]
f = sys.stdout
for xp in (numpy, cupy):
print(xp.__name__)
for shape in shapes:
a = xp.ones(shape)
f.write('shape={:<15} => '.format(str(shape)))
try:
b = xp.nonzero(a)
f.write('{}\n'.format(b))
except Exception as e:
f.write('FAIL: {}\n'.format(e))
# get stack trace
cupy.nonzero(cupy.ones((0,)))
Result:
numpy
shape=() => (array([0]),)
shape=(0,) => (array([], dtype=int64),)
shape=(1,) => (array([0]),)
shape=(0, 2) => (array([], dtype=int64), array([], dtype=int64))
shape=(0, 0, 2, 0) => (array([], dtype=int64), array([], dtype=int64), array([], dtype=int64), array([], dtype=int64))
cupy
shape=() => (array([0]),)
shape=(0,) => FAIL: CUDA_ERROR_INVALID_VALUE: invalid argument
shape=(1,) => (array([0]),)
shape=(0, 2) => FAIL: CUDA_ERROR_INVALID_VALUE: invalid argument
shape=(0, 0, 2, 0) => FAIL: CUDA_ERROR_INVALID_VALUE: invalid argument
Traceback (most recent call last):
File "test-nonzero.py", line 26, in <module>
cupy.nonzero(cupy.ones((0,)))
File "/niboshi/repos/cupy/cupy/sorting/search.py", line 72, in nonzero
return a.nonzero()
File "cupy/core/core.pyx", line 810, in cupy.core.core.ndarray.nonzero (cupy/core/core.cpp:16210)
scan_index = scan(condition.astype(dtype).ravel())
File "cupy/core/core.pyx", line 3883, in cupy.core.core.scan (cupy/core/core.cpp:83826)
kern_scan(grid=((a.size - 1) // (2 * block_size) + 1,),
File "cupy/cuda/function.pyx", line 118, in cupy.cuda.function.Function.__call__ (cupy/cuda/function.cpp:3794)
_launch(
File "cupy/cuda/function.pyx", line 100, in cupy.cuda.function._launch (cupy/cuda/function.cpp:3431)
driver.launchKernel(
File "cupy/cuda/driver.pyx", line 170, in cupy.cuda.driver.launchKernel (cupy/cuda/driver.cpp:3262)
check_status(status)
File "cupy/cuda/driver.pyx", line 70, in cupy.cuda.driver.check_status (cupy/cuda/driver.cpp:1481)
raise CUDADriverError(status)
cupy.cuda.driver.CUDADriverError: CUDA_ERROR_INVALID_VALUE: invalid argument
CuPy version: latest master(v2.0.0a1)
Currently ufuncs defined in cupy.fusion accepts numpy arrays. This is not intended.
Expected behavior (cupy.math.arithmetic.add)
>>> cupy.math.arithmetic.add(numpy.array(1), numpy.array(2))
Traceback (most recent call last):
File "<stdin>", line 1, in <module>
File "cupy/core/elementwise.pxi", line 736, in cupy.core.core.ufunc.__call__ (cupy/core/core.cpp:48698)
args = _preprocess_args(args)
File "cupy/core/elementwise.pxi", line 110, in cupy.core.core._preprocess_args (cupy/core/core.cpp:36647)
raise TypeError('Unsupported type %s' % type(arg))
TypeError: Unsupported type <class 'numpy.ndarray'>
Fusion (cupy.add = cupy.fusion.add)
>>> cupy.fusion.add(numpy.array(1), numpy.array(2))
3
These ufuncs should accept numpy.ndarray only within @fuse decorator.
It seems like the biggest opportunity for this project is not to be used in new, small projects that are just getting off the ground, but rather to be used as a NumPy replacement for the many large projects that already rely on NumPy.
It would be really useful for those projects to have in-depth documentation on what they'll have to change to migrate from NumPy to CuPy. Ideally this would include:
Can't wait to see this gain wider adoption! GPU-speed machine learning algorithms sounds like a dream.
Using cupy (commit: 5062f61065caecb8b3910c452f51b1307f5d8121 on Windows 10) in my program, I got the following many error messages:
Traceback (most recent call last):
File "cupy\cuda\memory.pyx", line 358, in cupy.cuda.memory.PooledMemory.free
TypeError: 'NoneType' object is not callable
Exception ignored in: 'cupy.cuda.memory.PooledMemory.__dealloc__'
Traceback (most recent call last):
File "cupy\cuda\memory.pyx", line 358, in cupy.cuda.memory.PooledMemory.free
TypeError: 'NoneType' object is not callable
...
Traceback (most recent call last):
File "cupy\cuda\runtime.pyx", line 222, in cupy.cuda.runtime.free
File "cupy\cuda\runtime.pyx", line 130, in cupy.cuda.runtime.check_status
cupy.cuda.runtime.CUDARuntimeError: cudaErrorInvalidDevicePointer: invalid device pointer
Exception ignored in: 'cupy.cuda.memory.Memory.__dealloc__'
...
Traceback (most recent call last):
File "xxx\__init__.py", line xxx, in xxx
return float(xpy.sum((y - z)**2, dtype=xpy.float64))
File "cupy\core\core.pyx", line 1475, in cupy.core.core.ndarray.__float__
File "cupy\core\core.pyx", line 1531, in cupy.core.core.ndarray.get
File "cupy\cuda\memory.pyx", line 254, in cupy.cuda.memory.MemoryPointer.copy_to_host
File "cupy\cuda\runtime.pyx", line 241, in cupy.cuda.runtime.memcpy
File "cupy\cuda\runtime.pyx", line 130, in cupy.cuda.runtime.check_status
cupy.cuda.runtime.CUDARuntimeError: cudaErrorIllegalAddress: an illegal memory access was encountered
Traceback (most recent call last):
File "cupy\cuda\memory.pyx", line 360, in cupy.cuda.memory.PooledMemory.free
AttributeError: 'weakref' object has no attribute 'cline_in_traceback'
Exception ignored in: 'cupy.cuda.memory.PooledMemory.__dealloc__'
Traceback (most recent call last):
File "cupy\cuda\memory.pyx", line 360, in cupy.cuda.memory.PooledMemory.free
AttributeError: 'weakref' object has no attribute 'cline_in_traceback'
Traceback (most recent call last):
File "cupy\cuda\runtime.pyx", line 222, in cupy.cuda.runtime.free
AttributeError: 'weakref' object has no attribute 'cline_in_traceback'
Exception ignored in: 'cupy.cuda.memory.Memory.__dealloc__'
Traceback (most recent call last):
File "cupy\cuda\runtime.pyx", line 222, in cupy.cuda.runtime.free
AttributeError: 'weakref' object has no attribute 'cline_in_traceback'
These errors only happen when I enable memory pool as the following code:
cupy.cuda.set_allocator(cupy.cuda.MemoryPool().malloc)I think the failure point (cupy.sum) is irreverent to the cause of these errors.
I have no details about these errors.
Is there any way to debug these errors more deeply?
On increasing unit size or batch size on chainer, users would meet cudaErrorMemoryAllocation at
Line 377 in 72c6d14
On such situation, I've heard from my collegue that users tune the unit size or batch size to be smaller to avoid cudaErrorMemoryAllocation, but they are unsure how much they must make these sizes be smaller.
I've heard that logging the allocation size on cudaErrorMemoryAllocation occuring is helpful for the tuning.
Does CuPy have a function equivalent to chainer.cuda.get_array_module ? I think it should be in CuPy because some users want to write CPU/GPU agnostic code with CuPy only.
Both README.md and Installation Guide include how to install CuPy, but they are inconsistent in at least
CUDA_PATH, whereas the former not.PATH and LD_LIBRARY_PATH to enable CUDA, whereas the latter not.It took me a lot of clicking to find the working docs just now.
Clicking on the badge link in the README takes me to a page that throws this error:
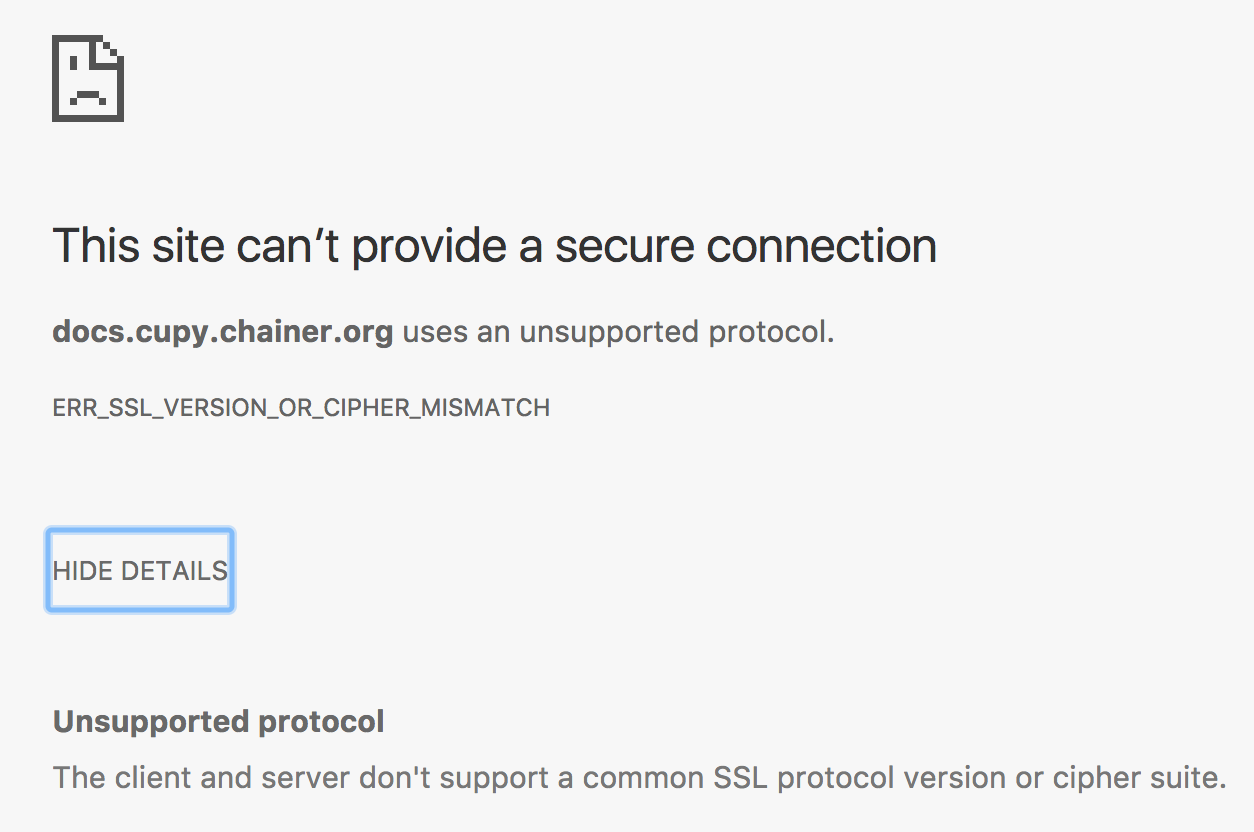
Clicking the top Google search link gives me the same error:
https://docs.cupy.chainer.org/
I found a working link a few results further down:
http://docs.chainer.org/en/stable/_modules/chainer/cuda.html
But then when I click "Edit on GitHub" on those docs, that link takes me to a GitHub page that doesn't exist, or doesn't have public permissions:
https://github.com/pfnet/chainer/blob/2a452f331c6d2634a05a4e3d9bee6ba5dbdd21f6/docs/source/_modules/chainer/cuda.rst
Quite excited to check out this project in more depth at some point!
Howdy!
I'm currently trying to add CuPy support to one of my packages, and it would be very useful if there was a cython level interface. Since I'd mostly like to replace BLAS with GPU support, I was wondering if there were any plans to support a cython level API similar to the API that scipy supports for BLAS. Thanks!
Code
import cupy
a = cupy.array([1, 2, 3])
cupy.array(a)
Log
Traceback (most recent call last):
File "test.py", line 4, in <module>
cupy.array(a)
File "/home/delta/dev/cupy/cupy/creation/from_data.py", line 26, in array
return core.array(obj, dtype, copy, ndmin)
File "cupy/core/core.pyx", line 1883, in cupy.core.core.array (cupy/core/core.cpp:59840)
File "cupy/core/core.pyx", line 1890, in cupy.core.core.array (cupy/core/core.cpp:59303)
File "cupy/core/core.pyx", line 263, in cupy.core.core.ndarray.astype (cupy/core/core.cpp:9428)
File "cupy/core/core.pyx", line 300, in cupy.core.core.ndarray.astype (cupy/core/core.cpp:8505)
TypeError: order not understood
This is because copy option of cupy.array is mistakenly interpreted as order option in cupy.ndarray here.
The current README asks to upgrade setuptools if the user uses an old one, but I would say this statement is too weak. Lazy users may (I mean, users are always lazy) just give a glance to this sentence and ignore it.
I guess a large portion of installation errors reported in Chainer v1 repository that say "cupy is not correctly installed" are due to an old setuptools.
I suggest to make the sentence much more strong, such as "You MUST upgrade setuptools before installing cupy", or maybe add a FAQ section (or a page?) to tell people that the very first thing to try after an installation failure is to upgrade it.
The cudnn.py file doesn't have comments hence it is very difficult to understand and use it. So can you please provide me a simple hello world example on how to use cupy's cudnn interface.
When using CuPy with memory pool from multi-threaded app, sometimes it fails to launch a kernel (CUDADriverError: CUDA_ERROR_INVALID_CONTEXT: invalid device context). I think this is because CUDA Driver API (to launch kernel) is called without establishing context on the host thread.
Here is a simple code to reproduce:
import chainer # Enable memory pool; without this line the issue does not reproduce.
import cupy
import threading
def run(size):
# Uncomment the following line to explicitly establish CUDA context
# on the current host thread:
#cupy.cuda.runtime.free(0)
print(cupy.arange(size, dtype=int))
size = 1024
# Run in main thread; this is OK.
# CuPy mallocs memory via Runtime API, then launches kernel with Driver API.
run(size)
# Run in another thread; this fails.
# The executed thread tries to launch kernel without establishing context,
# as Runtime API is not used (memory block acquired in the previous run is
# reused from pool.)
t = threading.Thread(target=run, args=(size,))
t.start()
t.join()As commented in the above code, I could workaround the problem by calling harmless Runtime API, e.g., cupy.cuda.runtime.free(0) to explicitly establish context on the host thread.
It would be great if CuPy could take care of such use case, but documenting the behavior may be enough.
Some tests which rely on random numbers are non-deterministic.
As the number of such test cases increases, the possibility of failure increases exponentially, even if each possibility for a single test is very low.
The CuPy contribution guide is almost copied from the one from Chainer v1. We should update so that it is consistent with Chainer v2. Currently the contribution guide of Chainer is being reviewed as chainer/chainer#2773 . We will port and modify it to be accommodated to CuPy.
We have to port chainer/chainer#2999 to CuPy.
Hi, I tried installing cupy and got an error like this.
File "/usr/lib64/python3.4/distutils/unixccompiler.py", line 87, in _fix_lib_args
runtime_library_dirs)
File "/usr/lib64/python3.4/distutils/unixccompiler.py", line 87, in _fix_lib_args
runtime_library_dirs)
File "/usr/lib64/python3.4/distutils/unixccompiler.py", line 86, in _fix_lib_args
self.__class__, self)._fix_lib_args(libraries, library_dirs,
RuntimeError: maximum recursion depth exceeded while calling a Python object
The env is: cupy-1.0.0.1 + Python 3.4.3 + Fedora23
$ cat /proc/version
Linux version 4.8.13-100.fc23.x86_64 ([email protected]) (gcc version 5.3.1 20160406 (Red Hat 5.3.1-6) (GCC) ) #1 SMP Fri Dec 9 14:51:40 UTC 2016
The full error log is this.
error.txt
A quick dirty workaround is to put something like this on _UnixCCompiler in "cupy_setup_build.py".
def _fix_lib_args(self, libraries, library_dirs, runtime_library_dirs):
"""Remove standard library path from rpath"""
libraries, library_dirs, runtime_library_dirs = super(
unixccompiler.UnixCCompiler, self)._fix_lib_args(libraries, library_dirs,
runtime_library_dirs)
libdir = sysconfig.get_config_var('LIBDIR')
if runtime_library_dirs and (libdir in runtime_library_dirs):
runtime_library_dirs.remove(libdir)
return libraries, library_dirs, runtime_library_dirs
It's taken from python3.5/distutils/unixccompiler.py and modified a bit.
Although we have switched from NVCC to NVRTC to compile kernels, CuPy searches for NVCC modules in its installation in setup.py and related scripts. We need to update them and corresponding installation document.
import numpy
import cupy
xp = cupy
a = xp.ones((4,4), dtype=numpy.float64)
b = xp.ones((4,4), dtype=numpy.int8) * 2
cond = xp.asarray(
[[True, False, False, True],
[False, True, True, True],
[True, False, False, True],
[True, False, True, False]])
print('a')
print(a)
print('b')
print(b)
print('cond')
print(cond)
z = xp.where(cond, a, b)
print('z')
print(z)
Result:
a
[[ 1. 1. 1. 1.]
[ 1. 1. 1. 1.]
[ 1. 1. 1. 1.]
[ 1. 1. 1. 1.]]
b
[[2 2 2 2]
[2 2 2 2]
[2 2 2 2]
[2 2 2 2]]
cond
[[ True False False True]
[False True True True]
[ True False False True]
[ True False True False]]
z
[[ 0.95294118 1. 1. 0.95294118]
[ 1. 0.95294118 0.95294118 0.95294118]
[ 0.95294118 1. 1. 0.95294118]
[ 0.95294118 1. 0.95294118 1. ]]
Resulted z is sometimes like above, sometimes like below.
z
[[ 1. 1. 1. 1.]
[ 1. 1. 1. 1.]
[ 1. 1. 1. 1.]
[ 1. 1. 1. 1.]]
Expected value (with xp = numpy):
z
[[ 1. 2. 2. 1.]
[ 2. 1. 1. 1.]
[ 1. 2. 2. 1.]
[ 1. 2. 1. 2.]]
a and b are swapped, the result will be as expected (the same as numpy).cupy.sorting.search.where. So fusion is not the cause.CUDA version
$ nvcc --version
nvcc: NVIDIA (R) Cuda compiler driver
Copyright (c) 2005-2016 NVIDIA Corporation
Built on Sun_Sep__4_22:14:01_CDT_2016
Cuda compilation tools, release 8.0, V8.0.44
https://docs-cupy.chainer.org/en/stable/tutorial/basic.html
There is one empty code block blow the sentence ”””In the following code, cp is an abbreviation of cupy, as np is numpy as is customarily done:""". This is strange.
Further, I guess cp, cupy, etc. in the above sentence should be described with coding format, not a plain text.
A declarative, efficient, and flexible JavaScript library for building user interfaces.
🖖 Vue.js is a progressive, incrementally-adoptable JavaScript framework for building UI on the web.
TypeScript is a superset of JavaScript that compiles to clean JavaScript output.
An Open Source Machine Learning Framework for Everyone
The Web framework for perfectionists with deadlines.
A PHP framework for web artisans
Bring data to life with SVG, Canvas and HTML. 📊📈🎉
JavaScript (JS) is a lightweight interpreted programming language with first-class functions.
Some thing interesting about web. New door for the world.
A server is a program made to process requests and deliver data to clients.
Machine learning is a way of modeling and interpreting data that allows a piece of software to respond intelligently.
Some thing interesting about visualization, use data art
Some thing interesting about game, make everyone happy.
We are working to build community through open source technology. NB: members must have two-factor auth.
Open source projects and samples from Microsoft.
Google ❤️ Open Source for everyone.
Alibaba Open Source for everyone
Data-Driven Documents codes.
China tencent open source team.