Mapping Degeneration Meets Label Evolution: Learning Infrared Small Target Detection with Single Point Supervision
Pytorch implementation of our Label Evolution with Single Point Supervision (LESPS). [Paper] [Web]
News: We recommend our newly-released repository BasicIRSTD, an open-source and easy-to-use toolbox for infrared small target detction. [link]
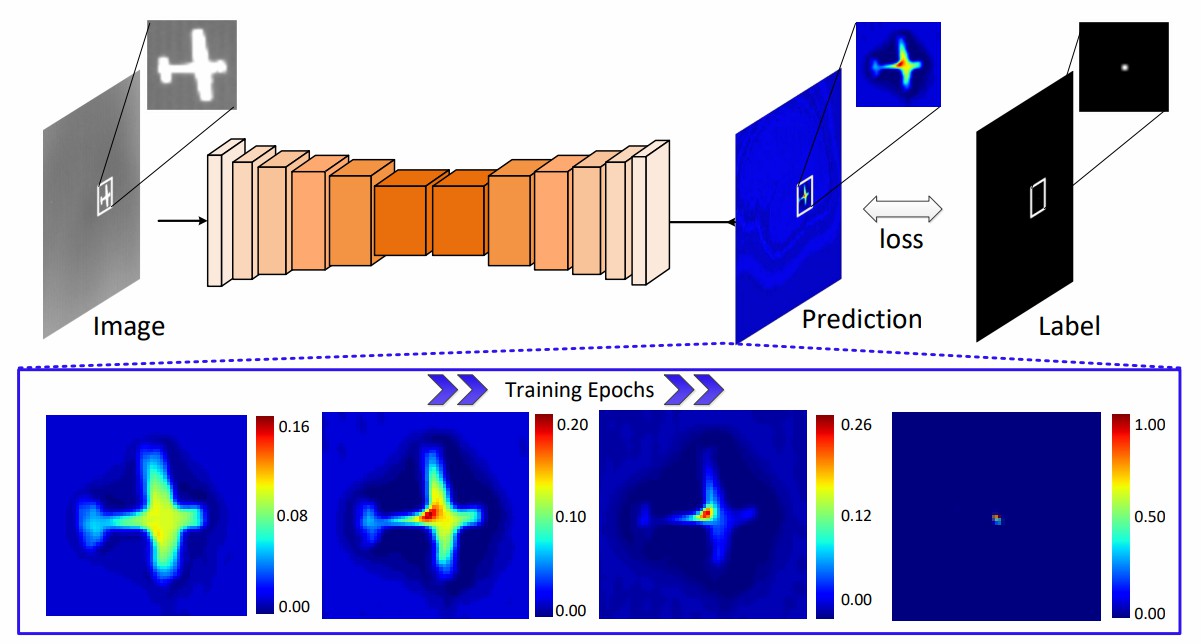
Fig. 1. Illustrations of mapping degeneration under point supervision. CNNs always tend to segment a cluster of pixels near the targets with low confidence at the early stage, and then gradually learn to predict groundtruth point labels with high confidence.
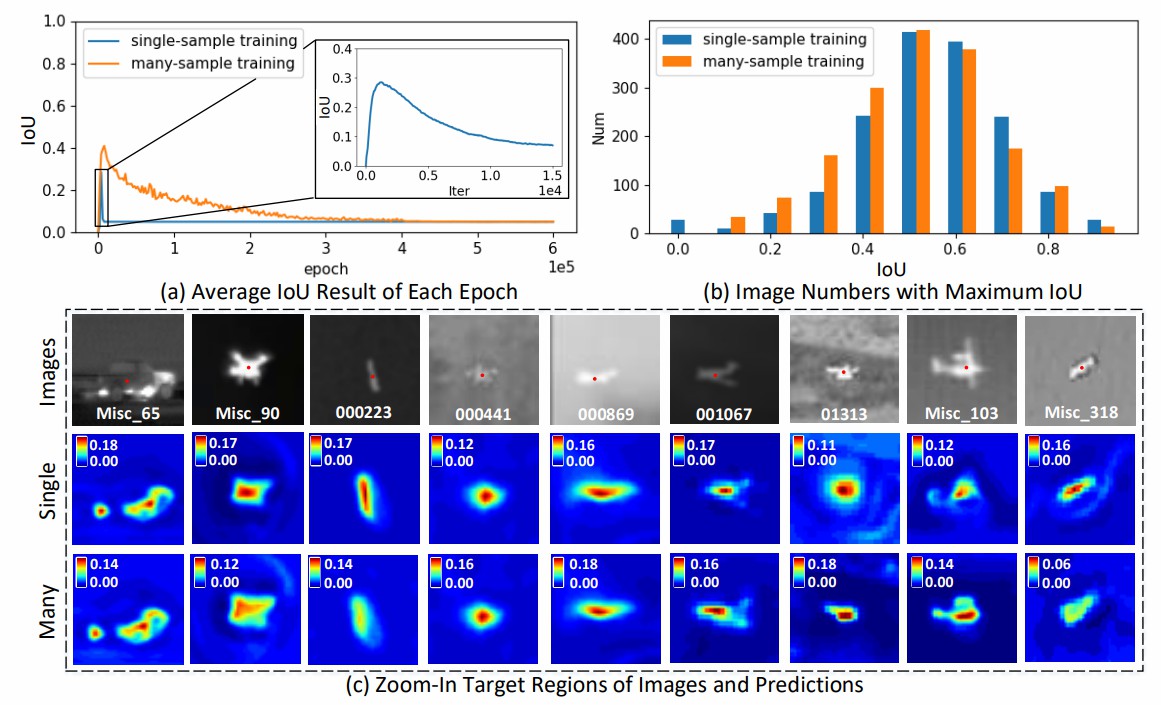
Fig. 2. Quantitative and qualitative illustrations of mapping
degeneration in CNNs.
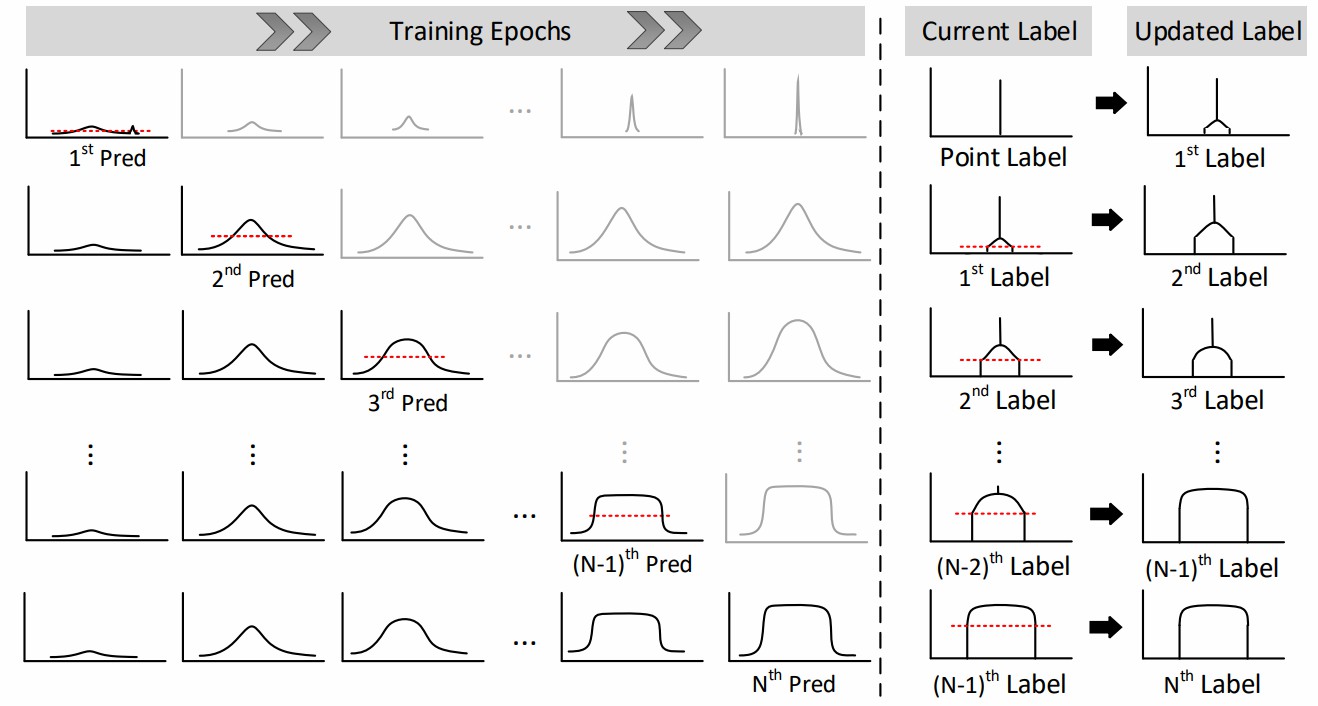
Fig. 3. Illustrations of Label Evolution with Single Point
Supervision (LESPS). During training, intermediate predictions of CNNs are used to progressively expand point labels to mask labels. Black arrows represent each round of label updates.
- Python 3
- pytorch (1.2.0), torchvision (0.4.0) or higher
- numpy, PIL
- NUAA-SIRST [download dir] [paper]
- NUDT-SIRST [download dir] [paper]
- IRSTD-1K [download dir] [paper]
SIRST3 is used for training, and is a combination of NUAA-SIRST, NUDT-SIRST, IRSTD-1K datasets.
Please first download datasets via Baidu Drive (key:1113), and place the datasets to the folder ./datasets/.
To gnenrate centroid annoation, run matlab code centroid_anno.m
To gnenrate coarse annoation, run matlab code coarse_anno.m
- Our project has the following structure:
├──./datasets/SIRST3/ │ ├── images │ │ ├── XDU0.png │ │ ├── Misc_1.png │ │ ├── ... │ │ ├── 001327.png │ ├── masks │ │ ├── XDU0.png │ │ ├── Misc_1.png │ │ ├── ... │ │ ├── 001327.png │ ├── masks_centroid │ │ ├── XDU0.png │ │ ├── Misc_1.png │ │ ├── ... │ │ ├── 001327.png │ ├── masks_coarse │ │ ├── XDU0.png │ │ ├── Misc_1.png │ │ ├── ... │ │ ├── 001327.png │ ├── img_idx │ │ ├── train_SIRST3.txt │ │ ├── test_SIRST3.txt │ │ ├── test_NUAA-SIRST.txt │ │ ├── test_NUDT-SIRST.txt │ │ ├── test_IRSTD-1K.txt
python train.py --model_names DNANet ALCNet ACM --dataset_names SIRST3 --label_type 'centroid'
python train.py --model_names DNANet ALCNet ACM --dataset_names SIRST3 --label_type 'coarse'python test.py --model_names DNANet ALCNet ACM --pth_dirs None --dataset_names NUAA-SIRST NUDT-SIRST IRSTD-1K
python test.py --model_names DNANet ALCNet ACM --pth_dirs SIRST3/DNANet_full.pth.tar SIRST3/DNANet_LESPS_centroid.pth.tar SIRST3/DNANet_LESPS_coarse.pth.tar SIRST3/ALCNet_full.pth.tar SIRST3/ALCNet_LESPS_centroid.pth.tar SIRST3/ALCNet_LESPS_coarse.pth.tar SIRST3/ACM_full.pth.tar SIRST3/ACM_LESPS_centroid.pth.tar SIRST3/ACM_LESPS_coarse.pth.tar --dataset_names NUAA-SIRST NUDT-SIRST IRSTD-1K
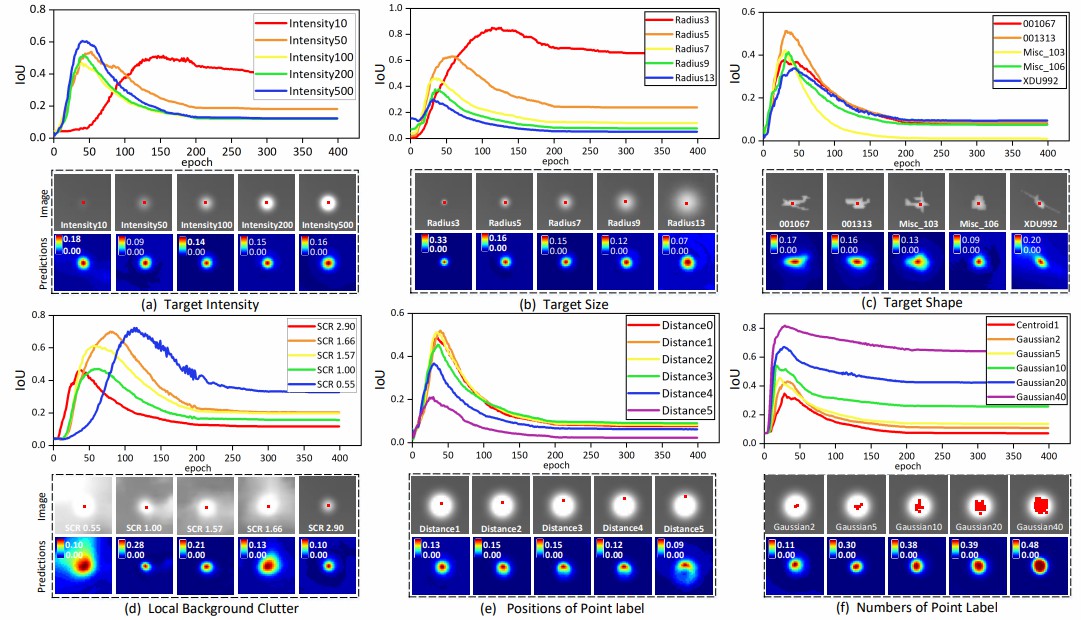
Fig. 4. IoU and visualize results of mapping degeneration with respect to different characteristics of targets (i.e.,(a) intensity, (b) size, (c) shape, and (d) local background clutter) and point labels (i.e.,(e) numbers and (f) locations). We visualize the zoom-in target regions of input images with GT point labels (i.e., red dots in images) and corresponding CNN predictions (in the epoch reaching maximum IoU).
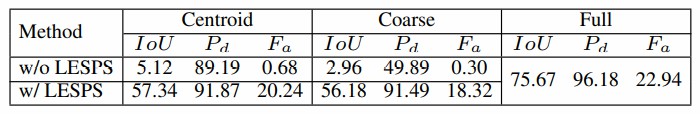
Table 1. Average results achieved by DNAnet with (w/) and without (w/o) LESPS under centroid, coarse point supervision together with full supervision.
Fig. 5. Quantitative and qualitative results of evolved target
masks.
Fig. 6. Visualizations of regressed labels during training and
network predictions during inference with centroid and coarse
point supervision.
Fig. 7. PA (P) and IoU (I) results of LESPS with respect to (a) initial evolution epoch, (b) Tb and (c) k of evolution threshold, and (d) evolution frequency.
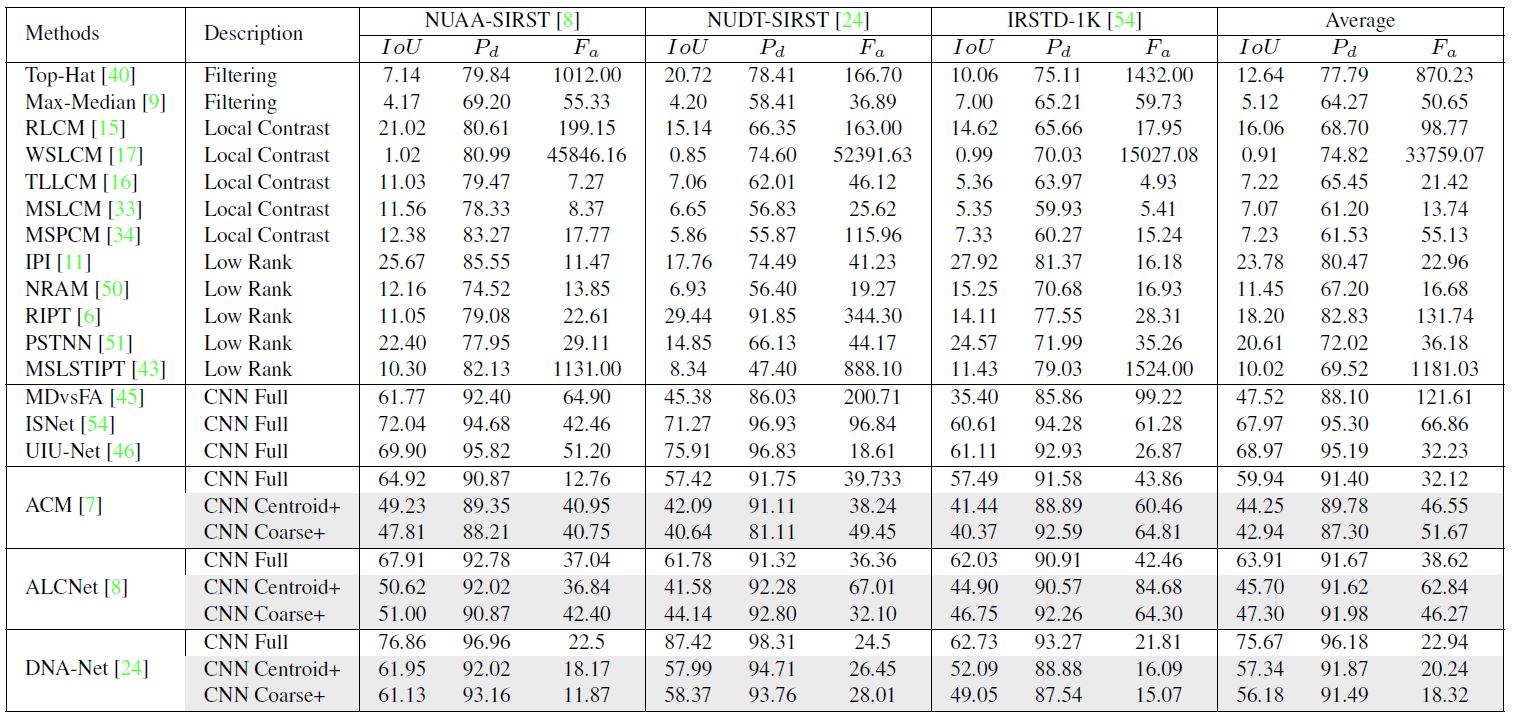
Table 2. Results of different methods. “CNN Full”, “CNN Centroid”, and “CNN Coarse” represent CNN-based methods under full supervision, centroid and coarse point supervision. “+” represents CNN-based methods equipped with LESPS.
Fig. 8. Visual detection results of different methods. Correctly detected targets and false alarms are highlighted by red and orange circles, respectively.
Table 3. Results of DNA-Net trained with pseudo labels generated by input intensity threshold, LCM-based methods and LESPS under centroid and coarse point supervision. Best results are shown in boldface.
@article{LESPS,
author = {Ying, Xinyi and Liu, Li and Wang, Yingqian and Li, Ruojing and Chen, Nuo and Lin, Zaiping and Sheng, Weidong and Zhou, Shilin},
title = {Mapping Degeneration Meets Label Evolution: Learning Infrared Small Target Detection with Single Point Supervision},
journal = {IEEE Conference on Computer Vision and Pattern Recognition (CVPR)},
year = {2023},
}
Welcome to raise issues or email to [email protected] for any question.