The RDML package is published in Oxford Bioinformatics:
Stefan Rödiger, Michał Burdukiewicz, Andrej-Nikolai Spiess, Konstantin Blagodatskikh; Enabling reproducible real-time quantitative PCR research: the RDML package, Bioinformatics, https://doi.org/10.1093/bioinformatics/btx528 (see also citation()).
Imports qPCR data from RDML v1.1 format files (Lefever et al. 2009) and transforms it to the appropriate format of the qpcR package (Ritz et al. 2008, Spiess et al. 2008) or chipPCR package. RDML (Real-time PCR Data Markup Language) is the recommended file format element in the Minimum Information for Publication of Quantitative Real-Time PCR Experiments (MIQE) guidelines (Bustin et al. 2009).
The stable version of the RDML package for R is hosted on CRAN and can be installed as any R package.
You can install the latest development version of the code using the devtools R package.
# Install devtools, if you haven't already.
install.packages("devtools")
library(devtools)
install_github("PCRuniversum/RDML")The manual is available online.
RDML imports various data formats (CSV, XMLX) besides the RDML format. Provided that the raw data
have a defined structure (as described in the vignette) the import should be
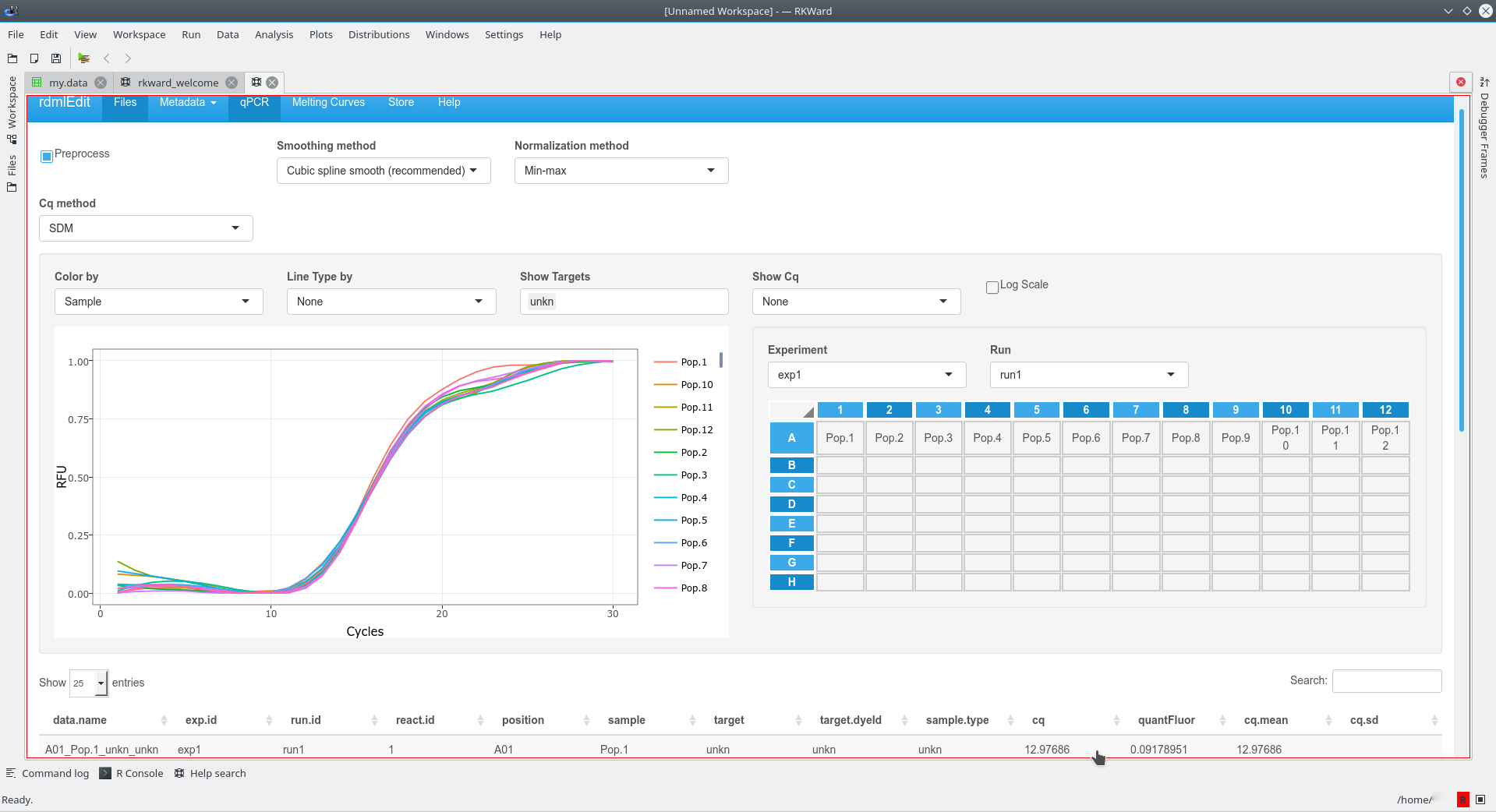
done by a few clicks. The example below shows the import of amplification curve
data, which were stored in a CSV file. The function rdmlEdit() was used in the
RKWard IDE/GUI for further processing. rdmlEdit may be also accessed as a web server (http://shtest.evrogen.net/rdmlEdit/).
Once imported enables rdmlEdit() and other functions from the RDML package complex
data visualization and processing in the R statistical computing environment.