Library for clinical NLP with spaCy.
MedSpaCy is currently in beta.
MedSpaCy is a library of tools for performing clinical NLP and text processing tasks with the popular spaCy
framework. The medspacy package brings together a number of other packages, each of which implements specific
functionality for common clinical text processing specific to the clinical domain, such as sentence segmentation,
contextual analysis and attribute assertion, and section detection.
medspacy is modularized so that each component can be used independently. All of medspacy is designed to be used
as part of a spacy processing pipeline. Each of the following modules is available as part of medspacy:
medspacy.preprocess: Destructive preprocessing for modifying clinical text before processingmedspacy.sentence_splitter: Clinical sentence segmentationmedspacy.ner: Utilities for extracting concepts from clinical textmedspacy.context: Implementation of the ConText for detecting semantic modifiers and attributes of entities, including negation and uncertaintymedspacy.section_detection: Clinical section detection and segmentationmedspacy.postprocess: Flexible framework for modifying and removing extracted entitiesmedspacy.io: Utilities for converting processed texts to structured data and interacting with databasesmedspacy.visualization: Utilities for visualizing concepts and relationships extracted from textSpacyQuickUMLS: UMLS concept extraction compatible with spacy and medspacy implemented by our fork of QuickUMLS. More detail on this component, how to use it, how to generate UMLS resources beyond the small UMLS sample can be found in this notebook.
Future work could include I/O, relations extraction, and pre-trained clinical models.
As of 10/2/2021 (version 0.2.0.0), medspaCy supports spaCy v3
You can install medspacy using setup.py:
python setup.py installOr with pip:
pip install medspacyTo install a previous version which uses spaCy 2:
pip install medspacy==medspacy 0.1.0.2The following packages are required and installed when medspacy is installed:
- spaCy v3
- pyrush
If you download other models, you can use them by providing the model itself or model name to medspacy.load(model_name):
import spacy; import medspacy
# Option 1: Load default
nlp = medspacy.load()
# Option 2: Load from existing model
nlp = spacy.load("en_core_web_sm", disable={"ner"})
nlp = medspacy.load(nlp)
# Option 3: Load from model name
nlp = medspacy.load("en_core_web_sm", disable={"ner"})Here is a simple example showing how to implement and visualize a simple rule-based pipeline using medspacy:
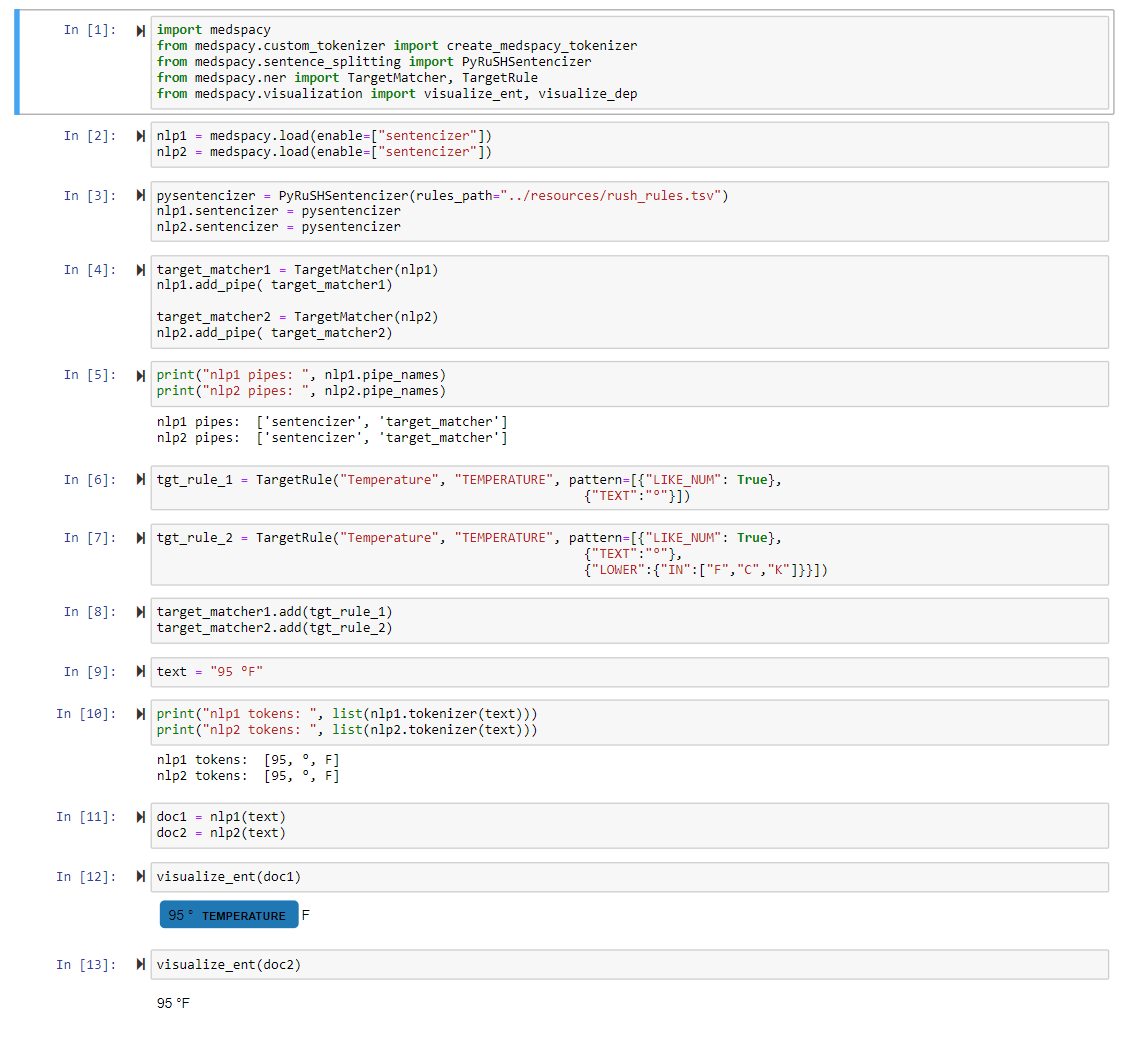
import medspacy
from medspacy.ner import TargetRule
from medspacy.visualization import visualize_ent
# Load medspacy model
nlp = medspacy.load()
print(nlp.pipe_names)
text = """
Past Medical History:
1. Atrial fibrillation
2. Type II Diabetes Mellitus
Assessment and Plan:
There is no evidence of pneumonia. Continue warfarin for Afib. Follow up for management of type 2 DM.
"""
# Add rules for target concept extraction
target_matcher = nlp.get_pipe("medspacy_target_matcher")
target_rules = [
TargetRule("atrial fibrillation", "PROBLEM"),
TargetRule("atrial fibrillation", "PROBLEM", pattern=[{"LOWER": "afib"}]),
TargetRule("pneumonia", "PROBLEM"),
TargetRule("Type II Diabetes Mellitus", "PROBLEM",
pattern=[
{"LOWER": "type"},
{"LOWER": {"IN": ["2", "ii", "two"]}},
{"LOWER": {"IN": ["dm", "diabetes"]}},
{"LOWER": "mellitus", "OP": "?"}
]),
TargetRule("warfarin", "MEDICATION")
]
target_matcher.add(target_rules)
doc = nlp(text)
visualize_ent(doc)For more detailed examples and explanations of each component, see the notebooks folder.
If you use medspaCy in your work, consider citing our paper! Presented at the AMIA Annual Symposium 2021, preprint available on Arxiv.
H. Eyre, A.B. Chapman, K.S. Peterson, J. Shi, P.R. Alba, M.M. Jones, T.L. Box, S.L. DuVall, O. V Patterson,
Launching into clinical space with medspaCy: a new clinical text processing toolkit in Python,
AMIA Annu. Symp. Proc. 2021 (in Press. (n.d.).
http://arxiv.org/abs/2106.07799.
@Article{medspacy,
Author="Eyre, H. and Chapman, A. B. and Peterson, K. S. and Shi, J. and Alba, P. R. and Jones, M. M. and Box, T. L. and DuVall, S. L. and Patterson, O. V. ",
Title="{{L}aunching into clinical space with medspa{C}y: a new clinical text processing toolkit in {P}ython}",
Journal="AMIA Annu Symp Proc",
Year="2021",
Volume="2021",
Pages="438--447"
}
}
Here are some links to projects or tutorials which use medSpacy. If you have a project which uses medSpaCy which you'd like to use, let us know!
- VA_COVID-19_NLP_BSV: An NLP pipeline for identifying positive cases of COVID-19 from clinical text. Deployed as part of the Department of Veterans Affairs response to COVID-19
- clinspacy: An R wrapper for spaCy, sciSpaCy, and medSpaCy for performing clinical NLP and UMLS linking in R
- mimic34md2020_materials: A crash course in clinical data science from the University of Melbourne. For medSpaCy materials, see
notebooks/nlp-*.ipynb - ReHouSED NLP: An NLP pipeline for studying Veteran housing stability and distinguishing between Veterans who are currently unstably housed and those who have exited homelessness