manuel-calzolari / sklearn-genetic Goto Github PK
View Code? Open in Web Editor NEWGenetic feature selection module for scikit-learn
Home Page: https://sklearn-genetic.readthedocs.io
License: GNU Lesser General Public License v3.0
Genetic feature selection module for scikit-learn
Home Page: https://sklearn-genetic.readthedocs.io
License: GNU Lesser General Public License v3.0
Please specify the parameter to be tuned.
Hi All,
Has anyone come across the issue described below? I'd appreciate any direction to help resolve this.
System information
OS Platform and Distribution: Windows 11 Home
Sklearn-genetic version: 0.5.1
deap version: 1.3.3
Scikit-learn version: 1.1.2
Python version: 3.8.13
Describe the bug
When running my pipeline to tune hyperparameters, this error occurs intermittently.
AssertionError: Assigned values have not the same length than fitness weights
I'm running TuneSearchCV (package tune-sklearn) to tune various hyperparameters of my pipeline below in this example, but have also encountered the error frequently when using GASearchCV (package sklearn-genetic-opt, also based on deap) :
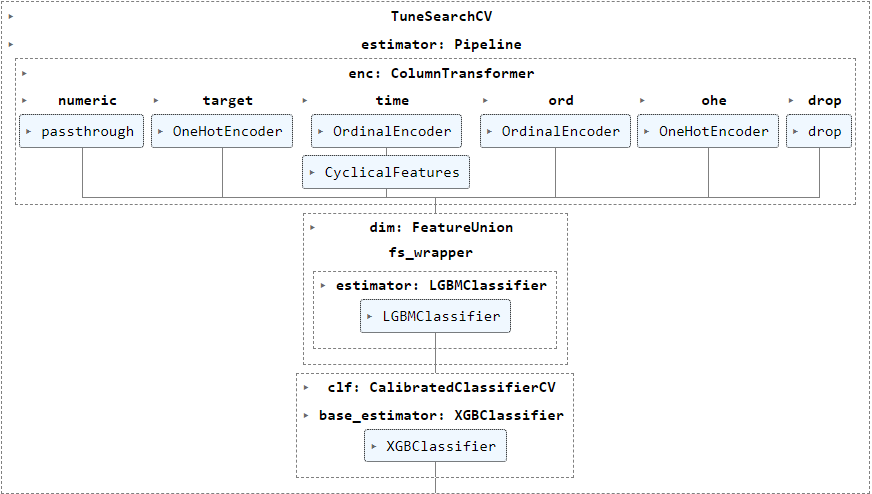
The following param_grid (generated using BayesSearchCV to show real information instead of the objects) show categorical values for various transformers steps enc__numeric, enc__target, enc__time__cyclicity and dim__fs_wrapper besides numerical parameter ranges for clf__base_estimator.
{'enc__numeric': Categorical(categories=('passthrough',
SmartCorrelatedSelection(selection_method='variance', threshold=0.9),
SmartCorrelatedSelection(cv='skf5',
estimator=LGBMClassifier(learning_rate=1.0,
max_depth=8,
min_child_samples=4,
min_split_gain=0.0031642299495941877,
n_jobs=1, num_leaves=59,
random_state=0, subsample=0.1,
verbose=-1),
scoring=make_scorer(average_precision_score, needs_proba=True, pos_label=1),
selection_method='model_performance', threshold=0.9)), prior=None),
'enc__target': Categorical(categories=(MeanEncoder(ignore_format=True), TargetEncoder(), MEstimateEncoder(),
WoEEncoder(ignore_format=True), PRatioEncoder(ignore_format=True),
BayesianTargetEncoder(columns=['Symbol', 'CandleType', 'h1CandleType1', 'h2CandleType1'],
prior_weight=3, suffix='')), prior=None),
'enc__time__cyclicity': Categorical(categories=(CyclicalFeatures(drop_original=True),
CycleTransformer(), RepeatingBasisFunction(n_periods=96)), prior=None),
'dim__fs_wrapper': Categorical(categories=('passthrough',
SelectFromModel(estimator=LGBMClassifier(learning_rate=1.0, max_depth=8,
min_child_samples=4,
min_split_gain=0.0031642299495941877,
n_jobs=1, num_leaves=59,
random_state=0, subsample=0.1,
verbose=-1),
importance_getter='feature_importances_'),
RFECV(cv=StratifiedKFold(n_splits=5, random_state=0, shuffle=True),
estimator=LGBMClassifier(learning_rate=1.0, max_depth=8,
min_child_samples=4,
min_split_gain=0.0031642299495941877, n_jobs=1,
num_leaves=59, random_state=0, subsample=0.1,
verbose=-1),
importance_getter='feature_importances_', min_features_to_select=10, n_jobs=1,
scoring=make_scorer(average_precision_score, needs_proba=True, pos_label=1), step=3),
GeneticSelectionCV(caching=True,
cv=StratifiedKFold(n_splits=5, random_state=0, shuffle=True),
estimator=LGBMClassifier(learning_rate=1.0, max_depth=8,
min_child_samples=4,
min_split_gain=0.0031642299495941877,
n_jobs=1, num_leaves=59,
random_state=0, subsample=0.1,
verbose=-1),
mutation_proba=0.1, n_gen_no_change=3, n_generations=20, n_population=50,
scoring=make_scorer(average_precision_score, needs_proba=True, pos_label=1))), prior=None),
'clf__base_estimator__eval_metric': Categorical(categories=('logloss', 'aucpr'), prior=None),
'clf__base_estimator__max_depth': Integer(low=2, high=8, prior='uniform', transform='identity'),
'clf__base_estimator__min_child_weight': Real(low=1e-05, high=1000, prior='log-uniform', transform='identity'),
'clf__base_estimator__colsample_bytree': Real(low=0.1, high=1.0, prior='uniform', transform='identity'),
'clf__base_estimator__subsample': Real(low=0.1, high=0.9999999999999999, prior='uniform', transform='identity'),
'clf__base_estimator__learning_rate': Real(low=1e-05, high=1, prior='log-uniform', transform='identity'),
'clf__base_estimator__gamma': Real(low=1e-06, high=1000, prior='log-uniform', transform='identity')}
To Reproduce
Steps to reproduce the behavior:
<<< Please let me know if you would like more information to reproduce the error >>>
Expected behavior
On occasions when it ran successfully, I got the following results for best_params_ as expected:
AssertionError Traceback (most recent call last)
Input In [130], in <cell line: 7>()
2 start = time()
3 # sv_results_ray = cross_val_clone(ray_pipe, X, y, cv_val[VAL], result_metrics, #scoring=score_metrics,
4 # return_estimator=True, return_train_score=True,
5 # optimise_threshold=True, granularity=THRESHOLD_GRAN
6 # )
----> 7 sv_results_ray = cross_val_thresh(ray_pipe, X, y, cv_val[VAL], result_metrics, #scoring=score_metrics,
8 return_estimator=True, return_train_score=True,
9 thresh_split=SPLIT,
10 )
11 end = time()Input In [46], in cross_val_thresh(estimator, X, y, cv, result_metrics, return_estimator, return_train_score, thresh_split, *args, **kwargs)
25 time_df.loc[i, 'split'] = i
27 start = time()
---> 28 est_i.fit(X_train, y_train, **kwargs) # **kwargs used to push callbacks to gen_pipe
29 end = time()
30 time_df.loc[i, 'fit_time'] = end - startFile C:\Anaconda3\envs\skl_py38\lib\site-packages\tune_sklearn\tune_basesearch.py:622, in TuneBaseSearchCV.fit(self, X, y, groups, tune_params, **fit_params)
597 def fit(self, X, y=None, groups=None, tune_params=None, **fit_params):
598 """Run fit with all sets of parameters.
599
600tune.runis used to perform the fit procedure.
(...)
620
621 """
--> 622 return self._fit(X, y, groups, tune_params, **fit_params)File C:\Anaconda3\envs\skl_py38\lib\site-packages\tune_sklearn\tune_basesearch.py:589, in TuneBaseSearchCV._fit(self, X, y, groups, tune_params, **fit_params)
587 refit_start_time = time.time()
588 if y is not None:
--> 589 self.best_estimator.fit(X, y, **fit_params)
590 else:
591 self.best_estimator.fit(X, **fit_params)File C:\Anaconda3\envs\skl_py38\lib\site-packages\sklearn\pipeline.py:378, in Pipeline.fit(self, X, y, **fit_params)
352 """Fit the model.
353
354 Fit all the transformers one after the other and transform the
(...)
375 Pipeline with fitted steps.
376 """
377 fit_params_steps = self._check_fit_params(**fit_params)
--> 378 Xt = self._fit(X, y, **fit_params_steps)
379 with _print_elapsed_time("Pipeline", self._log_message(len(self.steps) - 1)):
380 if self._final_estimator != "passthrough":File C:\Anaconda3\envs\skl_py38\lib\site-packages\sklearn\pipeline.py:336, in Pipeline._fit(self, X, y, **fit_params_steps)
334 cloned_transformer = clone(transformer)
335 # Fit or load from cache the current transformer
--> 336 X, fitted_transformer = fit_transform_one_cached(
337 cloned_transformer,
338 X,
339 y,
340 None,
341 message_clsname="Pipeline",
342 message=self._log_message(step_idx),
343 **fit_params_steps[name],
344 )
345 # Replace the transformer of the step with the fitted
346 # transformer. This is necessary when loading the transformer
347 # from the cache.
348 self.steps[step_idx] = (name, fitted_transformer)File C:\Anaconda3\envs\skl_py38\lib\site-packages\joblib\memory.py:594, in MemorizedFunc.call(self, *args, **kwargs)
593 def call(self, *args, **kwargs):
--> 594 return self._cached_call(args, kwargs)[0]File C:\Anaconda3\envs\skl_py38\lib\site-packages\joblib\memory.py:537, in MemorizedFunc._cached_call(self, args, kwargs, shelving)
534 must_call = True
536 if must_call:
--> 537 out, metadata = self.call(*args, **kwargs)
538 if self.mmap_mode is not None:
539 # Memmap the output at the first call to be consistent with
540 # later calls
541 if self._verbose:File C:\Anaconda3\envs\skl_py38\lib\site-packages\joblib\memory.py:779, in MemorizedFunc.call(self, *args, **kwargs)
777 if self._verbose > 0:
778 print(format_call(self.func, args, kwargs))
--> 779 output = self.func(*args, **kwargs)
780 self.store_backend.dump_item(
781 [func_id, args_id], output, verbose=self._verbose)
783 duration = time.time() - start_timeFile C:\Anaconda3\envs\skl_py38\lib\site-packages\sklearn\pipeline.py:870, in _fit_transform_one(transformer, X, y, weight, message_clsname, message, **fit_params)
868 with _print_elapsed_time(message_clsname, message):
869 if hasattr(transformer, "fit_transform"):
--> 870 res = transformer.fit_transform(X, y, **fit_params)
871 else:
872 res = transformer.fit(X, y, **fit_params).transform(X)File C:\Anaconda3\envs\skl_py38\lib\site-packages\sklearn\pipeline.py:1154, in FeatureUnion.fit_transform(self, X, y, **fit_params)
1133 def fit_transform(self, X, y=None, **fit_params):
1134 """Fit all transformers, transform the data and concatenate results.
1135
1136 Parameters
(...)
1152 sum ofn_components(output dimension) over transformers.
1153 """
-> 1154 results = self._parallel_func(X, y, fit_params, _fit_transform_one)
1155 if not results:
1156 # All transformers are None
1157 return np.zeros((X.shape[0], 0))File C:\Anaconda3\envs\skl_py38\lib\site-packages\sklearn\pipeline.py:1176, in FeatureUnion._parallel_func(self, X, y, fit_params, func)
1173 self._validate_transformer_weights()
1174 transformers = list(self._iter())
-> 1176 return Parallel(n_jobs=self.n_jobs)(
1177 delayed(func)(
1178 transformer,
1179 X,
1180 y,
1181 weight,
1182 message_clsname="FeatureUnion",
1183 message=self._log_message(name, idx, len(transformers)),
1184 **fit_params,
1185 )
1186 for idx, (name, transformer, weight) in enumerate(transformers, 1)
1187 )File C:\Anaconda3\envs\skl_py38\lib\site-packages\joblib\parallel.py:1043, in Parallel.call(self, iterable)
1034 try:
1035 # Only set self._iterating to True if at least a batch
1036 # was dispatched. In particular this covers the edge
(...)
1040 # was very quick and its callback already dispatched all the
1041 # remaining jobs.
1042 self._iterating = False
-> 1043 if self.dispatch_one_batch(iterator):
1044 self._iterating = self._original_iterator is not None
1046 while self.dispatch_one_batch(iterator):File C:\Anaconda3\envs\skl_py38\lib\site-packages\joblib\parallel.py:861, in Parallel.dispatch_one_batch(self, iterator)
859 return False
860 else:
--> 861 self._dispatch(tasks)
862 return TrueFile C:\Anaconda3\envs\skl_py38\lib\site-packages\joblib\parallel.py:779, in Parallel._dispatch(self, batch)
777 with self._lock:
778 job_idx = len(self._jobs)
--> 779 job = self._backend.apply_async(batch, callback=cb)
780 # A job can complete so quickly than its callback is
781 # called before we get here, causing self._jobs to
782 # grow. To ensure correct results ordering, .insert is
783 # used (rather than .append) in the following line
784 self._jobs.insert(job_idx, job)File C:\Anaconda3\envs\skl_py38\lib\site-packages\joblib_parallel_backends.py:208, in SequentialBackend.apply_async(self, func, callback)
206 def apply_async(self, func, callback=None):
207 """Schedule a func to be run"""
--> 208 result = ImmediateResult(func)
209 if callback:
210 callback(result)File C:\Anaconda3\envs\skl_py38\lib\site-packages\joblib_parallel_backends.py:572, in ImmediateResult.init(self, batch)
569 def init(self, batch):
570 # Don't delay the application, to avoid keeping the input
571 # arguments in memory
--> 572 self.results = batch()File C:\Anaconda3\envs\skl_py38\lib\site-packages\joblib\parallel.py:262, in BatchedCalls.call(self)
258 def call(self):
259 # Set the default nested backend to self._backend but do not set the
260 # change the default number of processes to -1
261 with parallel_backend(self._backend, n_jobs=self._n_jobs):
--> 262 return [func(*args, **kwargs)
263 for func, args, kwargs in self.items]File C:\Anaconda3\envs\skl_py38\lib\site-packages\joblib\parallel.py:262, in (.0)
258 def call(self):
259 # Set the default nested backend to self._backend but do not set the
260 # change the default number of processes to -1
261 with parallel_backend(self._backend, n_jobs=self._n_jobs):
--> 262 return [func(*args, **kwargs)
263 for func, args, kwargs in self.items]File C:\Anaconda3\envs\skl_py38\lib\site-packages\sklearn\utils\fixes.py:117, in _FuncWrapper.call(self, *args, **kwargs)
115 def call(self, *args, **kwargs):
116 with config_context(**self.config):
--> 117 return self.function(*args, **kwargs)File C:\Anaconda3\envs\skl_py38\lib\site-packages\sklearn\pipeline.py:870, in _fit_transform_one(transformer, X, y, weight, message_clsname, message, **fit_params)
868 with _print_elapsed_time(message_clsname, message):
869 if hasattr(transformer, "fit_transform"):
--> 870 res = transformer.fit_transform(X, y, **fit_params)
871 else:
872 res = transformer.fit(X, y, **fit_params).transform(X)File C:\Anaconda3\envs\skl_py38\lib\site-packages\sklearn\base.py:870, in TransformerMixin.fit_transform(self, X, y, **fit_params)
867 return self.fit(X, **fit_params).transform(X)
868 else:
869 # fit method of arity 2 (supervised transformation)
--> 870 return self.fit(X, y, **fit_params).transform(X)File C:\Anaconda3\envs\skl_py38\lib\site-packages\genetic_selection\gscv.py:279, in GeneticSelectionCV.fit(self, X, y, groups)
262 def fit(self, X, y, groups=None):
263 """Fit the GeneticSelectionCV model and then the underlying estimator on the selected
264 features.
265
(...)
277 instance (e.g.,GroupKFold).
278 """
--> 279 return self._fit(X, y, groups)File C:\Anaconda3\envs\skl_py38\lib\site-packages\genetic_selection\gscv.py:343, in GeneticSelectionCV._fit(self, X, y, groups)
340 print("Selecting features with genetic algorithm.")
342 with np.printoptions(precision=6, suppress=True, sign=" "):
--> 343 _, log = _eaFunction(pop, toolbox, cxpb=self.crossover_proba,
344 mutpb=self.mutation_proba, ngen=self.n_generations,
345 ngen_no_change=self.n_gen_no_change,
346 stats=stats, halloffame=hof, verbose=self.verbose)
347 if self.n_jobs != 1:
348 pool.close()File C:\Anaconda3\envs\skl_py38\lib\site-packages\genetic_selection\gscv.py:50, in _eaFunction(population, toolbox, cxpb, mutpb, ngen, ngen_no_change, stats, halloffame, verbose)
48 fitnesses = toolbox.map(toolbox.evaluate, invalid_ind)
49 for ind, fit in zip(invalid_ind, fitnesses):
---> 50 ind.fitness.values = fit
52 if halloffame is None:
53 raise ValueError("The 'halloffame' parameter should not be None.")File C:\Anaconda3\envs\skl_py38\lib\site-packages\deap\base.py:188, in Fitness.setValues(self, values)
187 def setValues(self, values):
--> 188 assert len(values) == len(self.weights), "Assigned values have not the same length than fitness weights"
189 try:
190 self.wvalues = tuple(map(mul, values, self.weights))AssertionError: Assigned values have not the same length than fitness weights
However, when I exclude dim__fs_wrapper from the pipeline, the error does not occur at all. The purpose of this transformer is to select a feature selection method from amongst 'passthrough' and estimators wrapped in SelectFromModel, RFECV and GeneticSelectionCV and the error seems to originate when GeneticSelectionCV is used for feature selection.
Additional context
Hello,
First of all, great work on creating this repository! Very lightweight and nicely coded. I've used it a lot for projects already, but lately I have been running into a strange problem...
I'm running the algorithm to tune for AUC, and the sum(selector.support_) is not equal to the number of features from the best individual in the logs that are printed to std output.
Selecting features with genetic algorithm.
gen nevals avg std min max
0 50 [ 0.62561904 225.48 ] [0.08697638 8.84135736] [ 0.52441945 207. ] [ 0.80103929 249. ]
1 36 [ 0.71277501 227.28 ] [0.06041145 8.97338286] [ 0.54962549 201. ] [ 0.81177172 249. ]
2 24 [ 0.75734538 228.3 ] [0.03028578 8.0628779 ] [ 0.67274129 201. ] [ 0.81177172 238. ]
3 32 [ 0.77257685 228.24 ] [0.03392161 8.02386441] [ 0.66479111 201. ] [ 0.84998056 241. ]
4 32 [ 0.7958877 229.84 ] [0.02928674 6.28445702] [ 0.70715193 217. ] [ 0.84998056 243. ]
5 38 [ 0.79477688 228.86 ] [0.07033557 6.90799537] [ 0.55381505 215. ] [ 0.85783745 244. ]
print(sum(selector.support_)) --> 231 while I expected 244 here.
Maybe something wrong with the HOF from DEAP?
Custom scorer using make_scorer causes GeneticSelectionCV to hang in Windows note this works fine in linux.
Is there any way to return multiple scores at the same time, and refer one metric as an estimator like GridSearchCV?
I am looking to use sklearn-genetic with a neural network, currently attempting to use with Keras NNs, although I am not necessarily tied to Keras.
I get the following error:
ValueError: Input 0 of layer sequential_2086 is incompatible with the layer: expected axis -1 of input shape to have value 180 but received input with shape (None, 118)
I understand why this is occurring - my NN input layer is expecting 180 features. Is there some way I can provide the number of features that sklearn-genetic is attempting to train with?
My KerasClassifier is defined as: estimator = KerasClassifier(lambda: create_nn_model(features=num_features), epochs=100) so I can dynamically supply this.
Can you suggest how I might use sklearn-genetic to select features for use in a NN?
Thanks for any help you can give.
I am playing around the repository and in order to have some kind of added value for my projects I think the project is missing one key thing. Validation on validation dataset. The GeneticSelectionCV will simply overfit on data and model on test sample that is taken apart will have nill performance.
Instead of
selector = selector.fit(
data.loc[train_mask, cols_pred],
data.loc[train_mask, col_target]
)The API should look something as follows
selector = selector.fit(
data.loc[train_mask, cols_pred],
data.loc[train_mask, col_target],
X_valid = data.loc[valid_mask, cols_pred],
y_valid = data.loc[valid_mask, col_target]
)What do you think?
I am having a difficult time understanding why estimator is needed in the first place.
Hey I have a dog breed dataset, shape (-1, 100,100,3). I get the following error
ValueError: Found array with dim 4. Estimator expected <= 2.
My estimator is my_model_2 a 5 layer CNN.
model = KerasClassifier(build_fn=my_model_2, epochs=10, validation_split = 0.1)
selector = GeneticSelectionCV(estimator=model,
cv=2,
verbose=1,
scoring="accuracy",
max_features=5,
n_population=50,
crossover_proba=0.5,
mutation_proba=0.2,
n_generations=40,
crossover_independent_proba=0.5,
mutation_independent_proba=0.05,
tournament_size=3,
n_gen_no_change=10,
caching=True,
n_jobs=-1)
the selector is default (except cv=2) for testing.
Is this supported? or do i need to change the shape of my data.
thank you.
is their any way to get the score of the each selected feature. need to plot a bargraph.
do you have example with complicated task when strong ML packages needed like lightGBM?
When Using a "Group" sklearn cross-validation object (in my case GroupShuffleSplit), the following error is raised
ValueError: The groups parameter should not be None
This issue might be related to #7646 of sklearn repo.
I managed to fix it by adding the groups parameter to the fit method, as done in other SearchCV algorithms (e.g. RandomizedSearchCV).
I can send a pull request if needed.
Dear Manuel
Congratulations on your code, and thank you very much for sharing it.
I'm using it for geological issues, and I would like to understand the independent crossover probability and also independent mutation probability. I was studying these topics in GA papers, but I didn't find a definition.
Can you explain it to me?
Best regards,
Michelle.
Hi, I am using the library for feature elimination
dataset: 400k * 50 all numeric columns
meta model :
RandomForestRegressor(bootstrap=True, criterion='mse',
n_estimators=25,
n_jobs = 8,
verbose=1)
algorithm setup:
GeneticSelectionCV(model,cv=3, verbose=1, n_population=30,
scoring=scoring,
max_features=40,
caching=True,
n_jobs=8)
I am getting a very long time of execution. It takes 1 or 2 hours of execution and still no result at all. Is it normal? How can I optimize?
is it possible to use F1 score for only positive label?
like
f1_score( y_test, current_prediction, pos_label = 'NO', average='binary')
nvm I looked at the wrong library
I came across some issues/warnings when using Python 3.8 and more recent versions of sklearn when debugging solutions to #11
For example, using Python 3.8 with scikit-learn 0.20.0 you get a couple issues:
imp instead of importlib/Users/john/.virtualenvs/sklearn-genetic/lib/python3.8/site-packages/sklearn/externals/joblib/externals/cloudpickle/cloudpickle.py:47: DeprecationWarning: the imp module is deprecated in favour of importlib; see the module's documentation for alternative uses
import imp
imp has been deprecated since Python 3.4 but I think the warning maybe only shows on Python 3.8? Relating the sklearn, this is well documented in scikit-learn/scikit-learn#12226 and scikit-learn/scikit-learn#12434 and solved I think in 0.20.1 onwards.
This is another incompatibility between Python 3.8 and the bundled versions of joblib/cloudpickle in sklearn 0.20.0, similar to this.
Full traceback:
In [6]: import sklearn
---------------------------------------------------------------------------
TypeError Traceback (most recent call last)
<ipython-input-6-b7c74cbf5af0> in <module>
----> 1 import sklearn
~/.virtualenvs/sklearn-genetic/lib/python3.8/site-packages/sklearn/__init__.py in <module>
63 from . import __check_build
64 from .base import clone
---> 65 from .utils._show_versions import show_versions
66
67 __check_build # avoid flakes unused variable error
~/.virtualenvs/sklearn-genetic/lib/python3.8/site-packages/sklearn/utils/__init__.py in <module>
11
12 from .murmurhash import murmurhash3_32
---> 13 from .validation import (as_float_array,
14 assert_all_finite,
15 check_random_state, column_or_1d, check_array,
~/.virtualenvs/sklearn-genetic/lib/python3.8/site-packages/sklearn/utils/validation.py in <module>
25 from ..exceptions import NotFittedError
26 from ..exceptions import DataConversionWarning
---> 27 from ..utils._joblib import Memory
28 from ..utils._joblib import __version__ as joblib_version
29
~/.virtualenvs/sklearn-genetic/lib/python3.8/site-packages/sklearn/utils/_joblib.py in <module>
16 from joblib import logger
17 else:
---> 18 from ..externals.joblib import __all__ # noqa
19 from ..externals.joblib import * # noqa
20 from ..externals.joblib import __version__ # noqa
~/.virtualenvs/sklearn-genetic/lib/python3.8/site-packages/sklearn/externals/joblib/__init__.py in <module>
117 from .numpy_pickle import load
118 from .compressor import register_compressor
--> 119 from .parallel import Parallel
120 from .parallel import delayed
121 from .parallel import cpu_count
~/.virtualenvs/sklearn-genetic/lib/python3.8/site-packages/sklearn/externals/joblib/parallel.py in <module>
30 LokyBackend)
31 from ._compat import _basestring
---> 32 from .externals.cloudpickle import dumps, loads
33 from .externals import loky
34
~/.virtualenvs/sklearn-genetic/lib/python3.8/site-packages/sklearn/externals/joblib/externals/cloudpickle/__init__.py in <module>
1 from __future__ import absolute_import
2
----> 3 from .cloudpickle import *
4
5 __version__ = '0.5.6'
~/.virtualenvs/sklearn-genetic/lib/python3.8/site-packages/sklearn/externals/joblib/externals/cloudpickle/cloudpickle.py in <module>
149
150
--> 151 _cell_set_template_code = _make_cell_set_template_code()
152
153
~/.virtualenvs/sklearn-genetic/lib/python3.8/site-packages/sklearn/externals/joblib/externals/cloudpickle/cloudpickle.py in _make_cell_set_template_code()
130 )
131 else:
--> 132 return types.CodeType(
133 co.co_argcount,
134 co.co_kwonlyargcount,
TypeError: an integer is required (got type bytes)Hi,
Thanks for such a nice implementation of GA for feature selection.
I am having an issue with passing the sample_weight fit parameter while using GeneticSelectionCV.
How this parameter should be passed while fitting GeneticSelectionCV?
Here are the details of my work:
The classifier I use is clf = XGBoostClassifier
For feature selection I use the following code where the rest of the parameters are the default ones:
genetic_CV = GeneticSelectionCV(estimator=clf)
Since I need to preprocess my data before fitting, I basically use the following pipeline for feature selection using GeneticSelectionCV:
ga_pipeline = Pipeline( steps=[('preprocessing',preprocessor), ('ga',genetic_CV)])
I think normally the fit parameter sample_weight should be passed like:
ga_pipeline.fit(X_train, y_train, ga__sample_weight=_weight_)
but it raises an error:
fit() got an unexpected keyword argument
sample_weight
Hi All,
Looks like SKL 1.3.0 may have broken things!
System information
OS Platform and Distribution: Windows 11 Pro
Sklearn-genetic version: 0.5.1
deap version: 1.4.1
Scikit-learn version: 1.3.0
Python version: 3.10.12
Describe the bug
Merely importing GeneticSelectionCV results in an error as below:
ImportError Traceback (most recent call last)
Cell In[102], line 1
----> 1 from genetic_selection import GeneticSelectionCVFile ~\Miniconda3\envs\skl310\lib\site-packages\genetic_selection_init_.py:16
1 # sklearn-genetic - Genetic feature selection module for scikit-learn
2 # Copyright (C) 2016-2022 Manuel Calzolari
3 #
(...)
13 # You should have received a copy of the GNU Lesser General Public License
14 # along with this program. If not, see http://www.gnu.org/licenses/.
---> 16 from .gscv import GeneticSelectionCV
18 version = '0.5.1'
20 all = ['GeneticSelectionCV']File ~\Miniconda3\envs\skl310\lib\site-packages\genetic_selection\gscv.py:22
20 import numpy as np
21 from sklearn.utils import check_X_y
---> 22 from sklearn.utils.metaestimators import if_delegate_has_method
23 from sklearn.base import BaseEstimator
24 from sklearn.base import MetaEstimatorMixinImportError: cannot import name 'if_delegate_has_method' from 'sklearn.utils.metaestimators' (C:\Users\naray\Miniconda3\envs\skl310\lib\site-packages\sklearn\utils\metaestimators.py)
To Reproduce
from genetic_selection import GeneticSelectionCV
Expected behaviour
The import shouldn't cause such an error.
Additional context
In jupyter notebook, I can run the following without issue:
estimator = KNeighborsClassifier(n_neighbors=16)
selector = GeneticSelectionCV(estimator,
cv=10,
verbose=1,
scoring="accuracy",
max_features=3,
n_population=1000,
crossover_proba=0.5,
mutation_proba=0.2,
n_generations=40,
crossover_independent_proba=0.5,
mutation_independent_proba=0.05,
tournament_size=3,
n_gen_no_change=10,
caching=True,
n_jobs=4)
selector = selector.fit(X, y)
However, as soon as I run it for a second time in the same ipython cell, all of the deap threads raise an exception. I've included the stack trace below.
Essentially, the above code can't run in a loop in ipython. Are there some threads which are not properly closed due to the interaction between GIL and ipython?
AttributeError: Can't get attribute 'FitnessMulti' on <module 'deap.creator' from 'D:\\anaconda\\envs\\a\\lib\\site-packages\\deap\\creator.py'>
AttributeError: Can't get attribute 'FitnessMulti' on <module 'deap.creator' from 'D:\\anaconda\\envs\\a\\lib\\site-packages\\deap\\creator.py'>
Process SpawnPoolWorker-58:
Traceback (most recent call last):
File "D:\anaconda\envs\a\lib\multiprocessing\process.py", line 297, in _bootstrap
self.run()
File "D:\anaconda\envs\a\lib\multiprocessing\process.py", line 99, in run
self._target(*self._args, **self._kwargs)
File "D:\anaconda\envs\a\lib\multiprocessing\pool.py", line 110, in worker
task = get()
File "D:\anaconda\envs\a\lib\multiprocessing\queues.py", line 354, in get
return _ForkingPickler.loads(res)
AttributeError: Can't get attribute 'FitnessMulti' on <module 'deap.creator' from 'D:\\anaconda\\envs\\a\\lib\\site-packages\\deap\\creator.py'>
Process SpawnPoolWorker-60:
Traceback (most recent call last):
File "D:\anaconda\envs\a\lib\multiprocessing\process.py", line 297, in _bootstrap
self.run()
File "D:\anaconda\envs\a\lib\multiprocessing\process.py", line 99, in run
self._target(*self._args, **self._kwargs)
File "D:\anaconda\envs\a\lib\multiprocessing\pool.py", line 110, in worker
task = get()
File "D:\anaconda\envs\a\lib\multiprocessing\queues.py", line 354, in get
return _ForkingPickler.loads(res)
AttributeError: Can't get attribute 'FitnessMulti' on <module 'deap.creator' from 'D:\\anaconda\\envs\\a\\lib\\site-packages\\deap\\creator.py'>
Environment Versions
Python 3.6.8pip 18.10.2Steps to replicate
pyenv shell 3.6.8 and mkvirtualenv sklearn-genetic-test to create fresh virtualenv.pip install sklearn-genetic==0.2from genetic_selection import GeneticSelectionCVExpected result
Running from genetic_selection import GeneticSelectionCV works without error.
Actual result
(sklearn-genetic-test) ➜ sklearn-genetic-test python
Python 3.6.8 (default, May 22 2020, 14:02:32)
[GCC 4.2.1 Compatible Apple LLVM 11.0.0 (clang-1100.0.33.17)] on darwin
Type "help", "copyright", "credits" or "license" for more information.
>>> from genetic_selection import GeneticSelectionCV
/Users/john/.virtualenvs/sklearn-genetic-test/lib/python3.6/site-packages/sklearn/utils/deprecation.py:143: FutureWarning: The sklearn.metrics.scorer module is deprecated in version 0.22 and will be removed in version 0.24. The corresponding classes / functions should instead be imported from sklearn.metrics. Anything that cannot be imported from sklearn.metrics is now part of the private API.
warnings.warn(message, FutureWarning)
/Users/john/.virtualenvs/sklearn-genetic-test/lib/python3.6/site-packages/sklearn/utils/deprecation.py:143: FutureWarning: The sklearn.feature_selection.base module is deprecated in version 0.22 and will be removed in version 0.24. The corresponding classes / functions should instead be imported from sklearn.feature_selection. Anything that cannot be imported from sklearn.feature_selection is now part of the private API.
warnings.warn(message, FutureWarning)
Traceback (most recent call last):
File "<stdin>", line 1, in <module>
File "/Users/john/.virtualenvs/sklearn-genetic-test/lib/python3.6/site-packages/genetic_selection/__init__.py", line 32, in <module>
from sklearn.externals.joblib import cpu_count
ModuleNotFoundError: No module named 'sklearn.externals.joblib'
>>> quit()
(sklearn-genetic-test) ➜ sklearn-genetic-test pip -V
pip 18.1 from /Users/john/.virtualenvs/sklearn-genetic-test/lib/python3.6/site-packages/pip (python 3.6)
(sklearn-genetic-test) ➜ sklearn-genetic-test python -c "from genetic_selection import GeneticSelectionCV"
/Users/john/.virtualenvs/sklearn-genetic-test/lib/python3.6/site-packages/sklearn/utils/deprecation.py:143: FutureWarning: The sklearn.metrics.scorer module is deprecated in version 0.22 and will be removed in version 0.24. The corresponding classes / functions should instead be imported from sklearn.metrics. Anything that cannot be imported from sklearn.metrics is now part of the private API.
warnings.warn(message, FutureWarning)
/Users/john/.virtualenvs/sklearn-genetic-test/lib/python3.6/site-packages/sklearn/utils/deprecation.py:143: FutureWarning: The sklearn.feature_selection.base module is deprecated in version 0.22 and will be removed in version 0.24. The corresponding classes / functions should instead be imported from sklearn.feature_selection. Anything that cannot be imported from sklearn.feature_selection is now part of the private API.
warnings.warn(message, FutureWarning)
Traceback (most recent call last):
File "<string>", line 1, in <module>
File "/Users/john/.virtualenvs/sklearn-genetic-test/lib/python3.6/site-packages/genetic_selection/__init__.py", line 32, in <module>
from sklearn.externals.joblib import cpu_count
ModuleNotFoundError: No module named 'sklearn.externals.joblib'
Hello, thanks for sharing your code.
I am using it on a dataset to distinguish cancer versus normal patterns from mass-spectrometric data.
I run the code and I am having a hard time understanding why there are two values of data on avg, std, min and max columns.
What does that mean? I am attaching a printscreen to illustrate.
Thanks.
Is this module limited to just classification? Can you perform regression with scoring set to rmse?
np.bool is now deprecated, instead numpy package only uses the built in bool python function. This means the package does not work anymore.
even after setting np.random.seed(xxx) the results are not predictable
Hello,
I have dependencies issues using the latest version of sklearn. I saw you addressed these in the latest commit 12ee9b2.
Could you bump a new version release so that I can properly lock the dependencies versions of my project ?
Many thanks for making this package available.
Hi,
below it is the definition of the selector class, n_jobs is by default set to 2, and two screen shot before and after running the code. It appears that n_jobs is running on all available cpus
class Selector:
def __init__(self, estimator=None, n_jobs: int = 2):
self.estimator = estimator
self.n_jobs = n_jobs
self.selector_model = None
self.select_estimator()
def select_estimator(self):
self.selector_model = GeneticSelectionCV(
self.estimator, cv=5, verbose=0,
scoring="f1_weighted", max_features=25,
n_population=50, crossover_proba=0.5,
mutation_proba=0.2, n_generations=50,
crossover_independent_proba=0.5,
mutation_independent_proba=0.04,
tournament_size=3, n_gen_no_change=10,
caching=True, n_jobs=self.n_jobs)
hello,Apply elitism operator to selected individuals in this repository??
Recently, scikit-learn has been updated to version 1.3.0 in Fedora Rawhide (development branch). Now tests for sklearn-genetic fail:
==================================== ERRORS ====================================
__________ ERROR collecting genetic_selection/tests/test_selection.py __________
ImportError while importing test module '/builddir/build/BUILD/sklearn-genetic-0.5.1/genetic_selection/tests/test_selection.py'.
Hint: make sure your test modules/packages have valid Python names.
Traceback:
/usr/lib64/python3.12/importlib/__init__.py:90: in import_module
return _bootstrap._gcd_import(name[level:], package, level)
genetic_selection/__init__.py:16: in <module>
from .gscv import GeneticSelectionCV
genetic_selection/gscv.py:22: in <module>
from sklearn.utils.metaestimators import if_delegate_has_method
E ImportError: cannot import name 'if_delegate_has_method' from 'sklearn.utils.metaestimators' (/usr/lib64/python3.12/site-packages/sklearn/utils/metaestimators.py)
=========================== short test summary info ============================
ERROR genetic_selection/tests/test_selection.py
!!!!!!!!!!!!!!!!!!!! Interrupted: 1 error during collection !!!!!!!!!!!!!!!!!!!!
=============================== 1 error in 0.64s ===============================
if_delegate_has_method has been deprecated since version 1.1 and is now (1.3.0) removed from metaestimators.py.
I tried a quick patch, but didn't succeed. I might look into it again, time permitting.
Sklearn-genetic-opt: https://github.com/rodrigo-arenas/Sklearn-genetic-opt
Is this package work only for classification or we can use it for regression models like Linear regression?
System information
OS Platform and Distribution: Windows 11 Pro
sklearn-genetic version: 0.5.1
Scikit-learn version: 1.3.1
Python version: 3.11.5
deap version: 1.4.1
numpy version: 1.24.3
Describe the bug
When trying to run
from genetic_selection import GeneticSelectionCV
it errors our as follows:
ImportError Traceback (most recent call last)
Cell In[105], line 13
7 from sklearn.feature_selection import (#VarianceThreshold, SelectKBest, SelectPercentile, GenericUnivariateSelect,
8 #f_classif, mutual_info_classif, chi2,
9 SelectFromModel,
10 RFECV, RFE,
11 )
12 from catboost import CatBoostClassifier
---> 13 from genetic_selection import GeneticSelectionCV
14 from sklearn_genetic import GAFeatureSelectionCV
15 #from boruta import BorutaPy
16 # from shapicant import PandasSelector
17 # import shap
File ~\Python\venvs\skl311\Lib\site-packages\genetic_selection\__init__.py:16
1 # sklearn-genetic - Genetic feature selection module for scikit-learn
2 # Copyright (C) 2016-2022 Manuel Calzolari
3 #
(...)
13 # You should have received a copy of the GNU Lesser General Public License
14 # along with this program. If not, see <http://www.gnu.org/licenses/>.
---> 16 from .gscv import GeneticSelectionCV
18 __version__ = '0.5.1'
20 __all__ = ['GeneticSelectionCV']
File ~\Python\venvs\skl311\Lib\site-packages\genetic_selection\gscv.py:22
20 import numpy as np
21 from sklearn.utils import check_X_y
---> 22 from sklearn.utils.metaestimators import if_delegate_has_method
23 from sklearn.base import BaseEstimator
24 from sklearn.base import MetaEstimatorMixin
ImportError: cannot import name 'if_delegate_has_method' from 'sklearn.utils.metaestimators' (C:\Users\naray\Python\venvs\skl311\Lib\site-packages\sklearn\utils\metaestimators.py)
Found this in the release notes of SKLearn 1.1.0
API Change utils.metaestimators.if_delegate_has_method is deprecated and will be removed in version 1.3. Use utils.metaestimators.available_if instead.
Might be one to fix along with #16 which is also driven by a SKLearn change.
Hello,
I tried to run the example in the README. Unfortunately, I got the following error:
multiprocessing.pool.RemoteTraceback:
"""
Traceback (most recent call last):
File "/home/g3rfx/anaconda3/envs/[redacted]/lib/python3.6/multiprocessing/pool.py", line 119, in worker
result = (True, func(*args, **kwds))
File "/home/g3rfx/anaconda3/envs/[redacted]/lib/python3.6/multiprocessing/pool.py", line 44, in mapstar
return list(map(*args))
File "/home/g3rfx/anaconda3/envs/[redacted]/lib/python3.6/site-packages/genetic_selection/gscv.py", line 134, in _evalFunction
scores_mean = np.mean(scores)
File "<__array_function__ internals>", line 6, in mean
File "/home/g3rfx/anaconda3/envs/[redacted]/lib/python3.6/site-packages/numpy/core/fromnumeric.py", line 3373, in mean
out=out, **kwargs)
File "/home/g3rfx/anaconda3/envs/[redacted]/lib/python3.6/site-packages/numpy/core/_methods.py", line 160, in _mean
ret = umr_sum(arr, axis, dtype, out, keepdims)
TypeError: unsupported operand type(s) for +: 'dict' and 'dict'
"""
The above exception was the direct cause of the following exception:
Traceback (most recent call last):
File "ga_feature_subset_selection.py", line 147, in <module>
ga_feature_selection()
File "ga_feature_subset_selection.py", line 141, in ga_feature_selection
selector.fit(X, y)
File "/home/g3rfx/anaconda3/envs/[redacted]/lib/python3.6/site-packages/genetic_selection/gscv.py", line 282, in fit
return self._fit(X, y)
File "/home/g3rfx/anaconda3/envs/[redacted]/lib/python3.6/site-packages/genetic_selection/gscv.py", line 346, in _fit
stats=stats, halloffame=hof, verbose=self.verbose)
File "/home/g3rfx/anaconda3/envs/[redacted]/lib/python3.6/site-packages/genetic_selection/gscv.py", line 55, in _eaFunction
fitnesses = toolbox.map(toolbox.evaluate, invalid_ind)
File "/home/g3rfx/anaconda3/envs/[redacted]/lib/python3.6/multiprocessing/pool.py", line 266, in map
return self._map_async(func, iterable, mapstar, chunksize).get()
File "/home/g3rfx/anaconda3/envs/[redacted]/lib/python3.6/multiprocessing/pool.py", line 644, in get
raise self._value
TypeError: unsupported operand type(s) for +: 'dict' and 'dict'
My environment is:
Ubuntu 20.04.2
Python 3.6.11
sklearn-genetic 0.3.0
scikit-learn 0.24.0
deap 1.3.1
Thank you in advance!
Hello,
First thank you for your work which works really great !
I am trying to compare results going with different optimization parameters for my model. I would like to store for each value of this parameter the computed maximum score value obtained with the variable selection. I understood that the feature selection is stored in selector.support_, but where is the score value (ex: accuracy) obtained with this feature selection ?
Thank you
First of, thanks for taking the time to creating this nice implementation a genetic feature selection module for scikit-learn.
My issue is that whenever I am trying to use it with a Pipeline in which I have a ColumnTransformer (with OneHotEncoder and StandardScaler) I get the following error: "Specifying the columns using strings is only supported for pandas DataFrames.". The error seems to originate from the return self.estimator_.fit(X[:, support_], y) call.
Any idea what may cause this? And is there a way for me to make my pipeline work with sklearn-genetic?
A declarative, efficient, and flexible JavaScript library for building user interfaces.
🖖 Vue.js is a progressive, incrementally-adoptable JavaScript framework for building UI on the web.
TypeScript is a superset of JavaScript that compiles to clean JavaScript output.
An Open Source Machine Learning Framework for Everyone
The Web framework for perfectionists with deadlines.
A PHP framework for web artisans
Bring data to life with SVG, Canvas and HTML. 📊📈🎉
JavaScript (JS) is a lightweight interpreted programming language with first-class functions.
Some thing interesting about web. New door for the world.
A server is a program made to process requests and deliver data to clients.
Machine learning is a way of modeling and interpreting data that allows a piece of software to respond intelligently.
Some thing interesting about visualization, use data art
Some thing interesting about game, make everyone happy.
We are working to build community through open source technology. NB: members must have two-factor auth.
Open source projects and samples from Microsoft.
Google ❤️ Open Source for everyone.
Alibaba Open Source for everyone
Data-Driven Documents codes.
China tencent open source team.