This is a collection of tools for studying parametric dependencies of black-box simulation codes or experiments and construction of reduced order response models over input parameter space.
proFit can be fed with a number of data points consisting of different input parameter combinations and the resulting output of the simulation under investigation. It then fits a response-surface through the point cloud using Gaussian process regression (GPR) models. This probabilistic response model allows to predict ("interpolate") the output at yet unexplored parameter combinations including uncertainty estimates. It can also tell you where to put more training points to gain maximum new information (experimental design) and automatically generate and start new simulation runs locally or on a cluster. Results can be explored and checked visually in a web frontend.
Telling proFit how to interact with your existing simulations is easy and requires no changes in your existing code. Current functionality covers starting simulations locally or on a cluster via Slurm, subsequent surrogate modelling using GPy, scikit-learn, as well as an active learning algorithm to iteratively sample at interesting points and a Markov-Chain-Monte-Carlo (MCMC) algorithm. The web frontend to interactively explore the point cloud and surrogate is based on plotly/dash.
- Compute evaluation points (e.g. from a random distribution) to run simulation
- Template replacement and automatic generation of run directories
- Starting parallel runs locally or on the cluster (SLURM)
- Collection of result output and postprocessing
- Response-model fitting using Gaussian Process Regression and Linear Regression
- Active learning to reduce number of samples needed
- MCMC to find a posterior parameter distribution (similar to active learning)
- Graphical user interface to explore the results
Currently, the code is under heavy development, so it should be cloned from GitHub via Git and pulled regularly.
sudo apt install python3-dev build-essentialTo enable compilation of the fortran modules the following is needed:
sudo apt install gfortran- numpy, scipy, matplotlib, sympy, pandas
- ChaosPy
- GPy
- scikit-learn
- h5py
- plotly/dash - for the UI
- ZeroMQ - for messaging
- sphinx - for documentation, only needed when
docsis specified - torch, GPyTorch - only needed when
gpuis specified
All dependencies are configured in setup.cfg and should be installed automatically when using pip.
Automatic tests use pytest.
To install proFit under Windows 10 we recommend using Windows Subsystem for Linux (WSL2) with the Ubuntu 20.04 LTS distribution (install guide).
After the installation of WSL2 execute the following steps in your Linux terminal (when asked press y to continue):
Make sure you have the right version of Python installed and the basic developer toolset available
sudo apt update
sudo apt install python3 python3-pip python3-dev build-essentialTo install proFit from Git (see below), make sure that the project is located in the Linux file system not the Windows system.
To configure the Python interpreter available in your Linux distribution in pycharm (tested with professional edition) follow this guide.
To install the latest stable version of proFit, use
pip install profitFor the latest pre-release, use
pip install --pre profitTo install proFit for the current user (--user) in development-mode (-e) use:
git clone https://github.com/redmod-team/profit.git
cd profit
pip install -e . --userCertain surrogates require a compiled Fortran backend. To enable compilation of the fortran modules during install:
USE_FORTRAN=1 pip install .
-
Make sure you have all the requirements mentioned above installed.
-
If
pipis not recognized try the following:
python3 -m pip install -e . --user- If pip warns you about PATH or proFit is not found close and reopen the terminal
and type
profit --helpto check if the installation was successful.
Install requirements for building the documentation using sphinx
pip install .[docs]
Additionally pandoc is required on a system level:
sudo apt install pandoc
Examples for different model codes are available under examples/:
fit: Simple fit via python interface.mockup: Simple model called by console command based on template directory.
Also, the integration tests under tests/integration_tests/ may be informative examples:
active_learning:- 1D: One dimensional mockup with active learning
- 2D: Two dimensional mockup with active learning
- Log: Active learning with logarithmic search space
- MCMC: Markov-Chain-Monte-Carlo application to mockup experimental data
mockup:- 1D
- 2D
- Custom postprocessor: Instead of the prebuilt postprocessor, a user-built class is used.
- Custom worker: A user-built worker function is used.
- Independent: Output with an independent (linear) variable additional to input parameters: f(t; u, v).
- KarhunenLoeve: Multi output surrogate model with Karhunen-Loeve encoder.
- Multi output: Multi output surrogate with two different output variables.
-
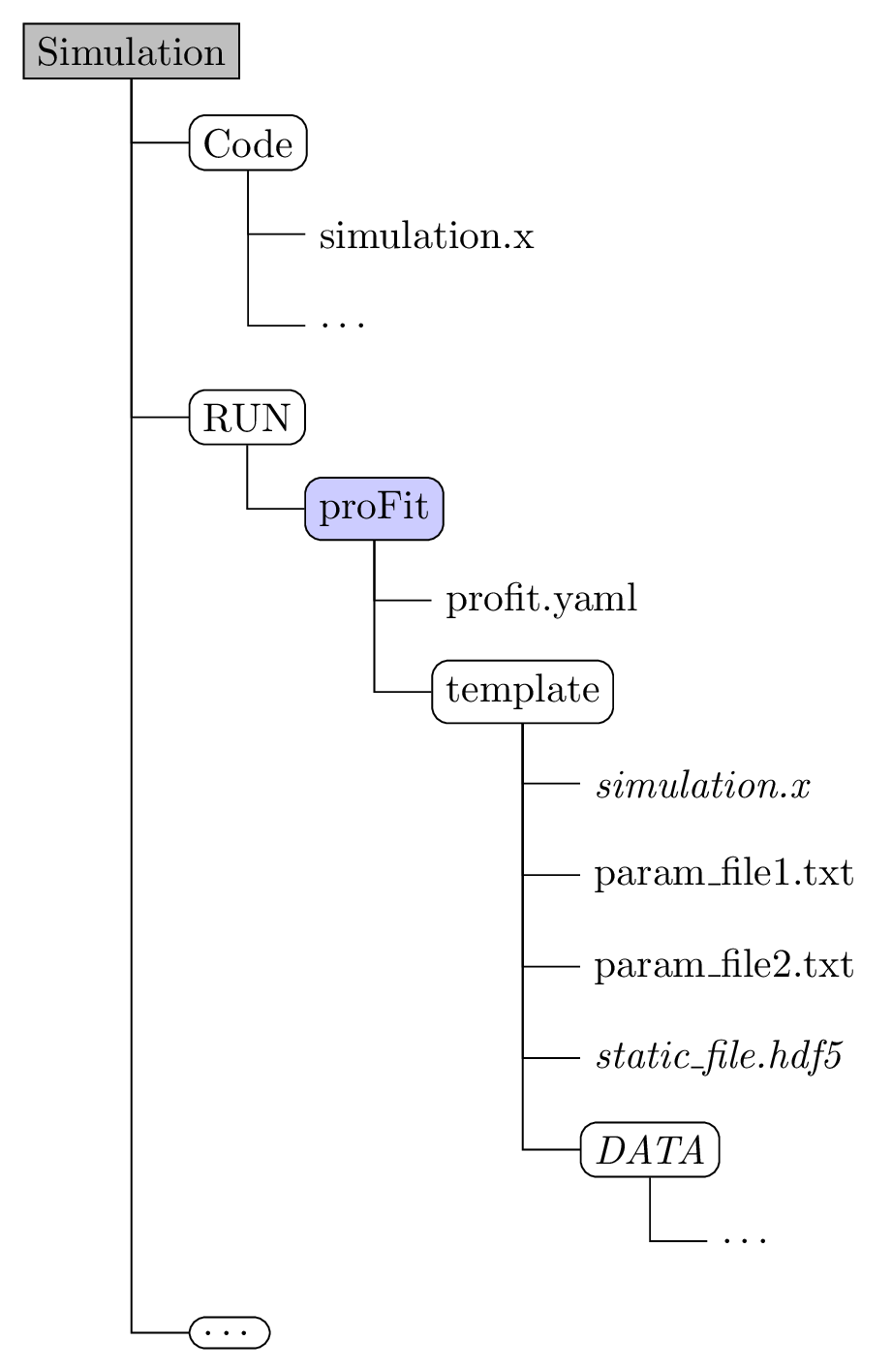
Create and enter a directory (e.g.
study) containingprofit.yamlfor your run. If your code is based on text configuration files for each run, copy the according directory totemplateand replace values of parameters to be varied within UQ/surrogate models by placeholders{param}. -
Running the simulations:
profit run
to start simulations at all the points. Per default the generated input variables are written to
input.txtand the output data is collected inoutput.txt.For each run of the simulation, proFit creates a run directory, fills the templates with the generated input data and collects the results. Each step can be customized with the configuration file.
-
To fit the model:
profit fit
Customization can be done with
profit.yamlagain. -
Explore data graphically:
profit ui
starts a Dash-based browser UI
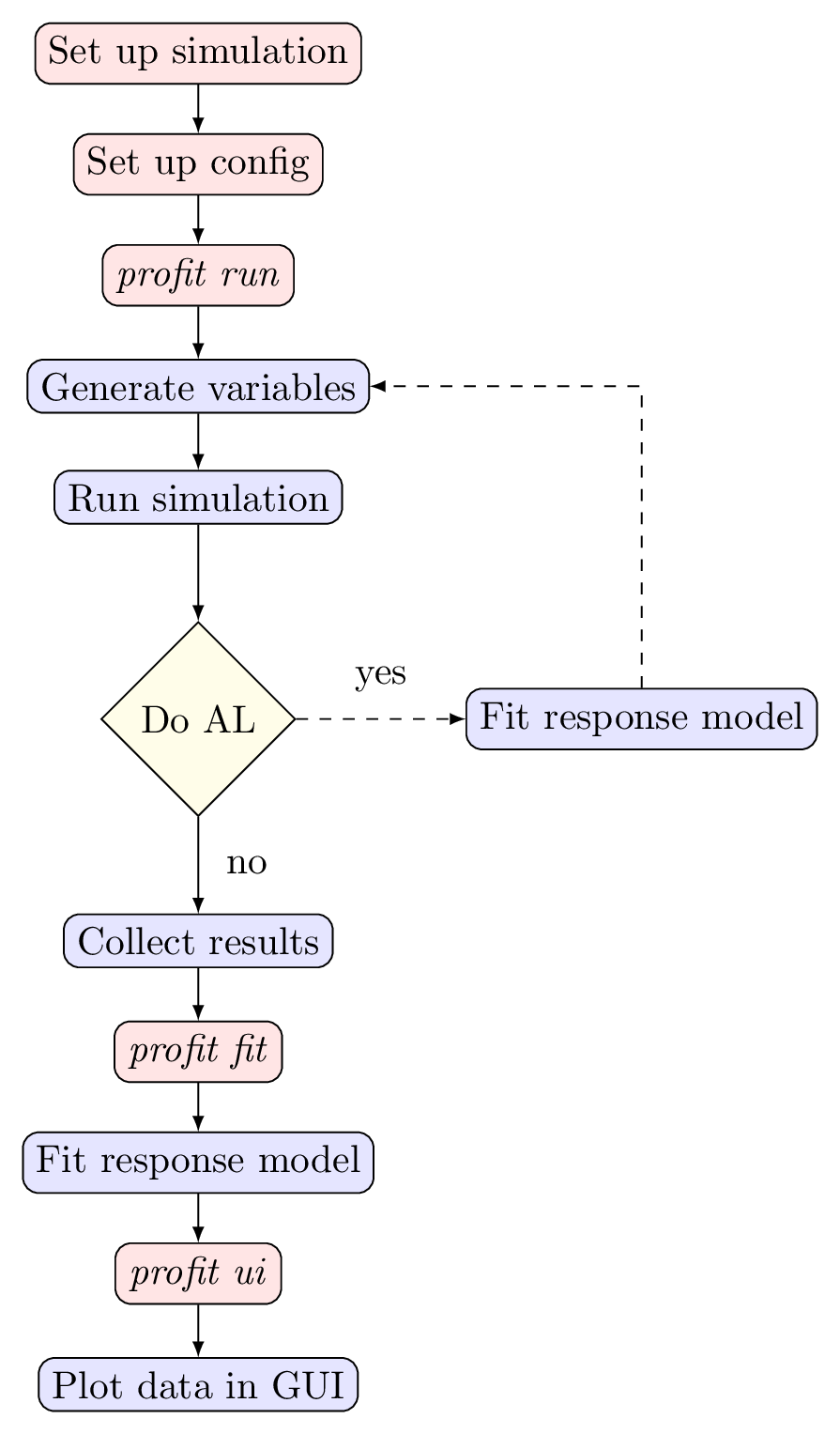
The figure below gives a graphical representation of the typical profit workflow described above. The boxes in red describe user actions while the boxes in blue are conducted by profit.
proFit supports scheduling the runs on a cluster using slurm. This is done entirely via the configuration files and the usage doesn't change.
profit ui starts a dash server and it is possible to remotely connect to it (e.g. via ssh port forwarding)
-
a configuration file: (default:
profit.yaml)- Add parameters and their distributions via
variables - Set paths and filenames
- Configure the run backend (how to interact with the simulation)
- Configure the fit / surrogate model
- Add parameters and their distributions via
-
the
templatedirectory- containing everything a simulation run needs (scripts, links to executables, input files, etc)
- input files use a template format where
{variable_name}is substituted with the generated values
-
a custom Postprocessor (optional)
- if the default postprocessors don't work with the simulation a custom one can be specified using the
includeparameter in the configuration.
- if the default postprocessors don't work with the simulation a custom one can be specified using the
Example directory structure: