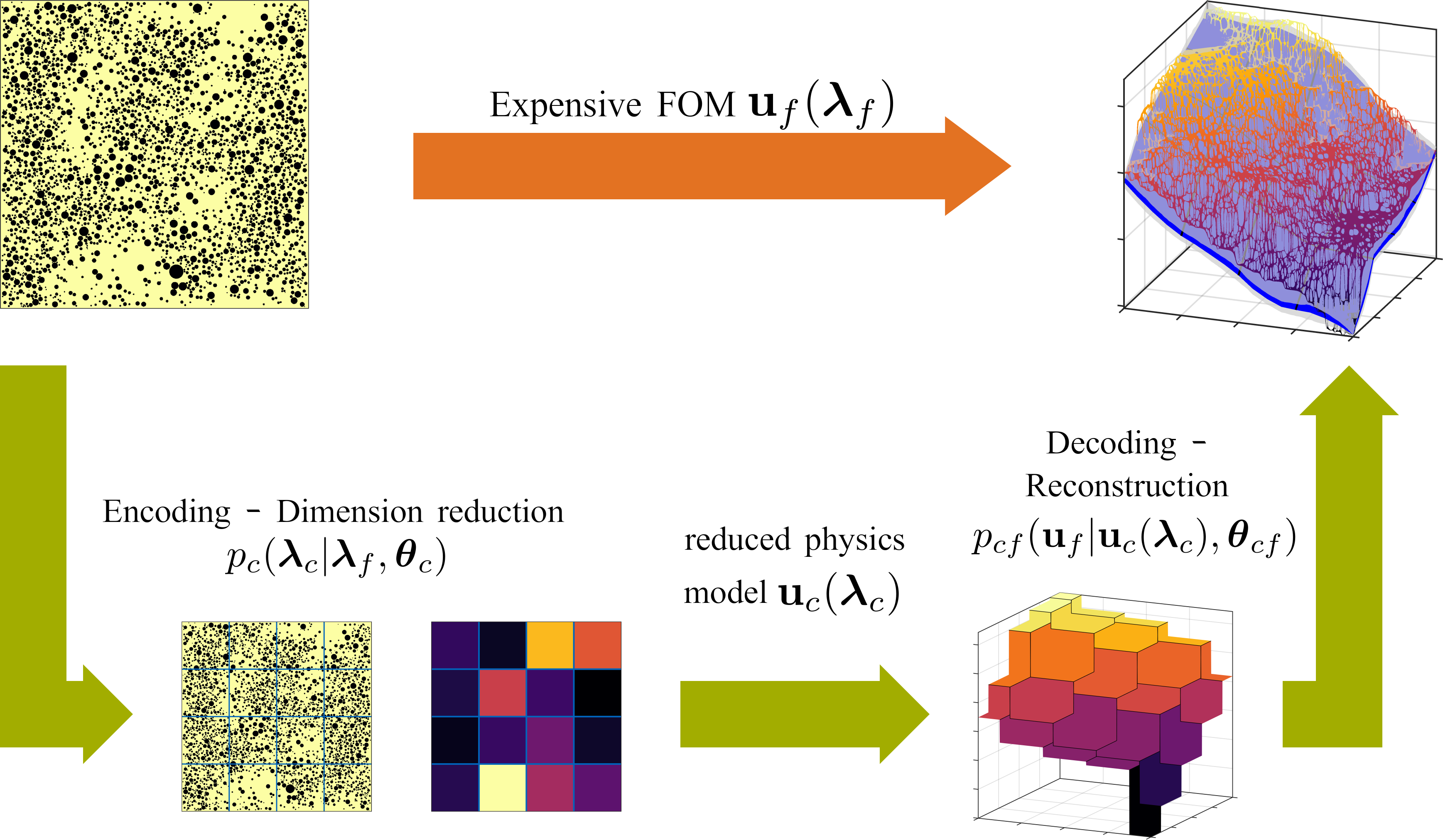
A physics-aware, probabilistic machine learning framework for coarse-graining high-dimensional systems in the Small Data regime
The automated construction of coarse-grained models represents a pivotal component in computer simulation of physical systems and is a key enabler in various analysis and design tasks related to uncertainty quantification. Pertinent methods are severely inhibited by the high-dimension of the parametric input and the limited number of training input/output pairs that can be generated when computationally demanding forward models are considered. Such cases are frequently encountered in the modeling of random heterogeneous media where the scale of the microstructure necessitates the use of high-dimensional random vectors and very fine discretizations of the governing equations. The present paper proposes a probabilistic Machine Learning framework that is capable of operating in the presence of Small Data by exploiting aspects of the physical structure of the problem as well as contextual knowledge. As a result, it can perform comparably well under extrapolative conditions. It unifies the tasks of dimensionality and model-order reduction through an encoder-decoder scheme that simultaneously identifies a sparse set of salient lower-dimensional microstructural features and calibrates an inexpensive, coarse-grained model which is predictive of the output. Information loss is accounted for and quantified in the form of probabilistic predictive estimates. The learning engine is based on Stochastic Variational Inference. We demonstrate how the variational objectives can be used not only to train the coarse-grained model, but also to suggest refinements that lead to improved predictions.
Schematic model overview.
- Matlab R2018b
- python 3.6
- FEniCS 2018.1.0
- mshr 2018.1.0
- scipy 1.1.0
- numpy 1.14.6
- Install Matlab 2018b, python 3.6 and all dependencies
- To clone this repo:
git clone https://github.com/congriUQ/physics_aware_surrogate.git
cd physics_aware_surrogate
Fine scale (Stokes flow) data is available.
1024 microstructures (center coordinates and radii of spherical exclusions, i.e. solid phase) as used in section 3.3 of the paper can be downloaded here.
1024 fine scale triangular meshes (corresponding to the microstructures above; vertex coordinates and cell connectivity) as used in section 3.3 of the paper can be downloaded here.
For correct usage, meshes and microstructures should both be saved under
/physics_aware_surrogate/data/meshSize=256/nonOverlappingDisks/margins=0.003_0.003_0.003_0.003/N~logn/mu=7.8/sigma=0.2/x~GP/cov=squaredExponential/l=0.08/sig_scale=1.2/r~lognGP/mu=-5.23/sigma=0.3/sigmaGP_r=0.4/l=0.05/
1024 solution fields (vertex values of pressure and velocity fields for meshes above) as used in section 3.3 of the paper can be downloaded here.
For correct usage, solutions should be saved under
/physics_aware_surrogate/data/meshSize=256/nonOverlappingDisks/margins=0.003_0.003_0.003_0.003/N~logn/mu=7.8/sigma=0.2/x~GP/cov=squaredExponential/l=0.08/sig_scale=1.2/r~lognGP/mu=-5.23/sigma=0.3/sigmaGP_r=0.4/l=0.05/p_bc=0.0/u_x=1.0-0.0x[1]_u_y=1.0-0.0x[0]/
To generate random microstructures as in sec. 3.3 of the paper, set up distribution parameters in lines 6-23 of ./genMicrostruct/genCorrMicrostruct.m as desired and run
/path/to/matlab -nodesktop -nodisplay -nosplash -r "addpath('./genMicrostruct') ; genCorrMicrostruct ; quit;"
.mat files containing the center coordinates and radii of each exclusion are stored in the folder ./data under a path name dependent on the microstructural parameters.
It is recommended to increase the stack size limit via
ulimit -s 32000
to avoid stack overflow during mesh generation.
Set up a conda Python 3 environment with the above modules (FEniCS, mshr, numpy, scipy). Activate the environment and run
python ./genMesh_cluster.py
This will generate 2-dimensional triangular meshes with random (approximately) circular exclusions based on the random microstructures saved in /path/to/microstructureInformationX.mat. The script will run until nMeshes meshes are generated.
Attention: mesh generation may take from a couple of minutes (for ~1000 circular exclusions) to several days (for ~10 000 exclusions or more). You may consider to run the mesh generation script on several CPUs in parallel. The script checks automatically to which microstructures there is no mesh/no currently running mesh generation and starts the one with the smallest number X. If you kill a mesh generation job for some reason, you should delete the file /path/to/computation_started.txt s.t. the above script can restart the generation of the corresponding meshes.
The mesh files are stored in the same folder as the microstructural data and the corresponding index, i.e. /path/to/meshX.mat belonging to the microstructure /path/to/microstructureInformationX.mat.
To generate fine scale Stokes flow PDE solutions, activate the FEniCS environment, set the microstructural parameters and the desired boundary conditions at the top of the script ./genSolution_cluster.py and run
python ./genSolution_cluster.py
The solutions are stored under path/to/p_bc=.../u_x=..._u_y=.../solutionX.mat, where p_bc, u_x, u_y encode the pressure and velocity boundary conditions.
Set the model parameters in the properties of the ModelParams class and the microstructure/fine scale data parameters in the StokesData class. In Matlab, run
>> trainModel
to train the surrogate according to section 2.4 of the paper. Convergence can be observed by the plots generated after every iteration as well as several parameters printed on the Matlab screen.
After fine scale data has been generated/downloaded and the model has been trained, converged model parameters are stored in the folder /physics_aware_surrogate/rom/data. The found parameters may be used to predict on independent test data. Run the script
>> predictionScript
to predict on a small test set. The code may be used to follow up on the paper results and to do own experiments.
If this code is relevant for your research, we would be grateful if you cite our preprint:
@article{Grigo2019,
title = "A physics-aware, probabilistic machine learning framework for coarse-graining high-dimensional systems in the Small Data regime",
journal = "Journal of Computational Physics",
volume = "397",
pages = "108842",
year = "2019",
issn = "0021-9991",
doi = "https://doi.org/10.1016/j.jcp.2019.05.053",
url = "http://www.sciencedirect.com/science/article/pii/S0021999119305261",
author = "Constantin Grigo and Phaedon-Stelios Koutsourelakis",
keywords = "Uncertainty quantification, Bayesian inference, Coarse-graining, Variational inference, Random media, Multi-physics models",
abstract = "The automated construction of coarse-grained models represents a pivotal component in computer simulation of physical systems and is a key enabler in various analysis and design tasks related to uncertainty quantification. Pertinent methods are severely inhibited by the high-dimension of the parametric input and the limited number of training input/output pairs that can be generated when computationally demanding forward models are considered. Such cases are frequently encountered in the modeling of random heterogeneous media where the scale of the microstructure necessitates the use of high-dimensional random vectors and very fine discretizations of the governing equations. The present paper proposes a probabilistic Machine Learning framework that is capable of operating in the presence of Small Data by exploiting aspects of the physical structure of the problem as well as contextual knowledge. As a result, it can perform comparably well under extrapolative conditions. It unifies the tasks of dimensionality and model-order reduction through an encoder-decoder scheme that simultaneously identifies a sparse set of salient lower-dimensional microstructural features and calibrates an inexpensive, coarse-grained model which is predictive of the output. Information loss is accounted for and quantified in the form of probabilistic predictive estimates. The learning engine is based on Stochastic Variational Inference. We demonstrate how the variational objectives can be used not only to train the coarse-grained model, but also to suggest refinements that lead to improved predictions."
}