blog's People
blog's Issues
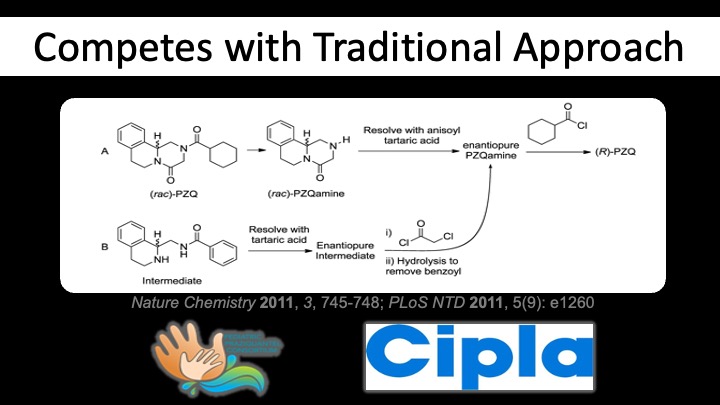
Praziquantel - The Development of an Open Science Asset
The first open source science project in which I was involved was one that derived an improved, realistic, inexpensive route to the active enantiomer of praziquantel. I've spoken about it many times over the years, and we published the science details and the open science mechanism. The project ran mainly 2009-2011, with most of the activity during a really fun sabbatical period I had in San Francisco Jan-June 2010. The point is that commercial racemic PZQ is super important as the medicine of choice for the neglected tropical disease schistosomiasis (Bilharzia), and also will be your cat/dog dewormer, but compliance is lessened because the medicine tastes terrible, and that taste arises mainly from the inactive enantiomer. So: make the active enantiomer, without the price going up, and we can improve the compliance while making the pill smaller.
The striking thing about the project was the speed and effectiveness of the open science. Highly qualified people contributed freely, mostly (about 75%) from the private sector, and accelerated the science to such an extent that we completed the project early. Many of the contributors were strangers to the core team, and I'm fairly sure that the contributions were made because the project was open. Again, the thing that is perhaps not understood (e.g., by folks who think that open science = just publishing open access) is that the contributions were made as the project was proceeding, and that those contributions changed the direction of the project. This is an essential feature of what I think of as open science. The possibility that others change what you're doing.
The other interesting feature was the way that our project was pitted against another that was working in secret within a contract research organisation. While I was initially miffed that we'd been secretly competing against another organisation (I think this was the devilish genius of our WHO collaborator, Piero Olliaro), the routes that emerged were not too dissimilar, and this acted as a kind of benchmarking exercise for an open team of mostly volunteers (30 or so people, in public) vs. a dedicated CRO (I think maybe 2 people full time, in secret). The CRO could see what we were doing, but we were unaware of the existence of the CRO - I think we were tipped off by a comment on Derek Lowe's blog.
Since then? Well, once we demonstrated that the asset, (R)-PZQ (it's being called arpraziquantel, doing away with the fiddly "(R)-"), could be made inexpensively and simply under ambient conditions on a gram scale, we were kind of done. The TDR division in WHO took what we'd achieved and we in the project kind of parted company with the downstream development, which is a logical thing to do. We were the science team, and we'd handed things off to those with expertise in public health and development.
The generics company Cipla were looking at scaling up what we'd found. I visited the site in Mumbai to speak to Yusuf Hamied about things and saw a truly beautiful kilo of enantiopure PZQ sitting there, and saw that one of the lead chemists was examining one of the TLCs from our open lab notebooks. Subsequently I was made aware of the Paediatric Praziquantel Consortium's work. It is they who are now really progressing things and doing wonderful work translating (R)-PZQ to widespread use. This has involved additional trials, e.g. to check that the single enantiomer is actually worth using - that the improvement in taste is worth it in terms of compliance.
And it was recently announced that Merck has signed a contract manufacturing agreement, which is wonderful news. I'm not sure of the route they're using (I've asked - a company in Nairobi is making it) but I like to think our public and heavily road-tested solution helped to move everything along in terms of feasibility, benchmarking etc.
Remember: this is an open asset - there is no IP protection, yet the molecule is progressing towards real use in the field and would positively impact public health for so many people. Throughout the project, which was a close alliance with WHO/TDR, we never had any doubt this would be a problem. There are other important examples of medicines that were brought through to patients unencumbered by protected IP/patents and which are helping people, like fexinidazole for sleeping sickness (in which Els Torreele played a major role) and ASAQ/Coarsucam for malaria. I so very much hope that (R)-PZQ joins them.
Open science can deliver medicines.
Hey Chemists! Planetary Forge Twitter Bot
I'm asking for advice, and potential solutions, for a twitter bot that is able to tweet out pictures of molecules to catch the eyes of chemists who may own related structures or know how to make them.
This is intended to contribute to solving the matchmaking problem of open source drug discovery: connecting projects that need molecules with those people (individuals or companies) who may be able to provide them.
Context
I gave a talk recently where I was describing how easy it is to miss relevant scientific work that is taking place elsewhere in the world (slide below). I was telling the story of how I had been scrolling through my Twitter feed and I'd screeched to a halt when I noticed a picture from one of the SGC's open lab notebooks. The structures were remarkably similar to structures we were looking at in OSM Series 3. One thing led to another and we and the SGC co-evaluated each others' molecules, but the point about the story is that I could so easily have missed it. I suspect I'm missing things all the time.
What we need is the magic bot that automatically connects people working in the open on similar molecules, SCINDR, that we've proposed (and just need 50K to implement). But we don't have that yet. Nor yet do we have a good Molecular Craigslist solution.
As I said in the talk "Finding people working on relevant things shouldn't rely on scrolling through pictures on Twitter". But then I added "although actually a bot that tweets out pictures of molecules isn't a terrible idea."
The need
We humans respond well to pictures, and we chemists respond particularly well to pictures of chemical structures. We understand them quickly and can place them in context well: Do I have that structure? Have I read about them recently or do I know someone looking at these? How would I make that?
In open source drug discovery projects we have openly available sheets of molecules that accumulate as the projects progress. These sheets contain SMILES/InChI strings. Here are examples from malaria, TB and mycetoma.
What we need is a bot that takes a random entry from one of these sheets, renders a nice chemical structure, and tweets out the picture of the molecule along with a link to where the interested person might find more info (e.g. the relevant project's landing page) as well as a chemical string (to make the tweet useful to other bots). So the idea is to attract the attention of human chemists, and make it easy for those interested people to connect to the project to see if they can help further.
This could be done with molecules that have been made already, or molecules that are needed next in a project.
Here's a mocked-up tweet, and I used the hashtag #planetaryforge to try to get across the underlying vision. It's a little like the Molecular Craigslist concept, but made specifically to catch the attention of human chemists.
The request
Anyone want to try this out by making a prototype bot? We can call it The Germinator.
Company Contributions to Open Science Projects
I talk about open science projects a lot, in particular ones that are aimed at the development of new medicines. A common question is "do companies contribute?"
The answer is yes. A lot. All the time.
This surprises many people, but shouldn't. I've written about this before, but wanted to flag it up again because contributions from companies happen all the time in open science projects.
I suspect this is well-known in software. Salaried company people giving time and expertise to open projects. The "Google 20%" - freedom to contribute to things that don't help the company's bottom line (and don't harm it, either). That's relevant, but not the whole story, since there are also huge open source projects that have stronger links to companies where the outputs help the company.
We first demonstrated this in an experimental science project with the transformative impact on the discovery of a simple route to the active form of an important drug. Spontaneous, high quality inputs from companies (both ideas and lab work) in a basic science project towards an IP-free medicine that is now looking like it is going to impact a great many people.
Over the last 15 years of doing open science medicinal chemistry projects, it's happened over and over and over and over. Small companies and big ones. Junior people and senior pharma professionals. Clear, public demonstrations of expertise by the company (that can lead to future investments - a successful funding round for Optibrium, for example, occurred after their successful participation in an Open Source Malaria competition). Good "unvarnished" PR. Fun and freedom for employees. Employee satisfaction at helping with something, e.g. in global health. Openness guaranteeing that the industry contribution makes a difference (i.e. it's needed), and does not disproportionately benefit a competitor. The Structural Genomics Consortium has seen the same thing over the years.
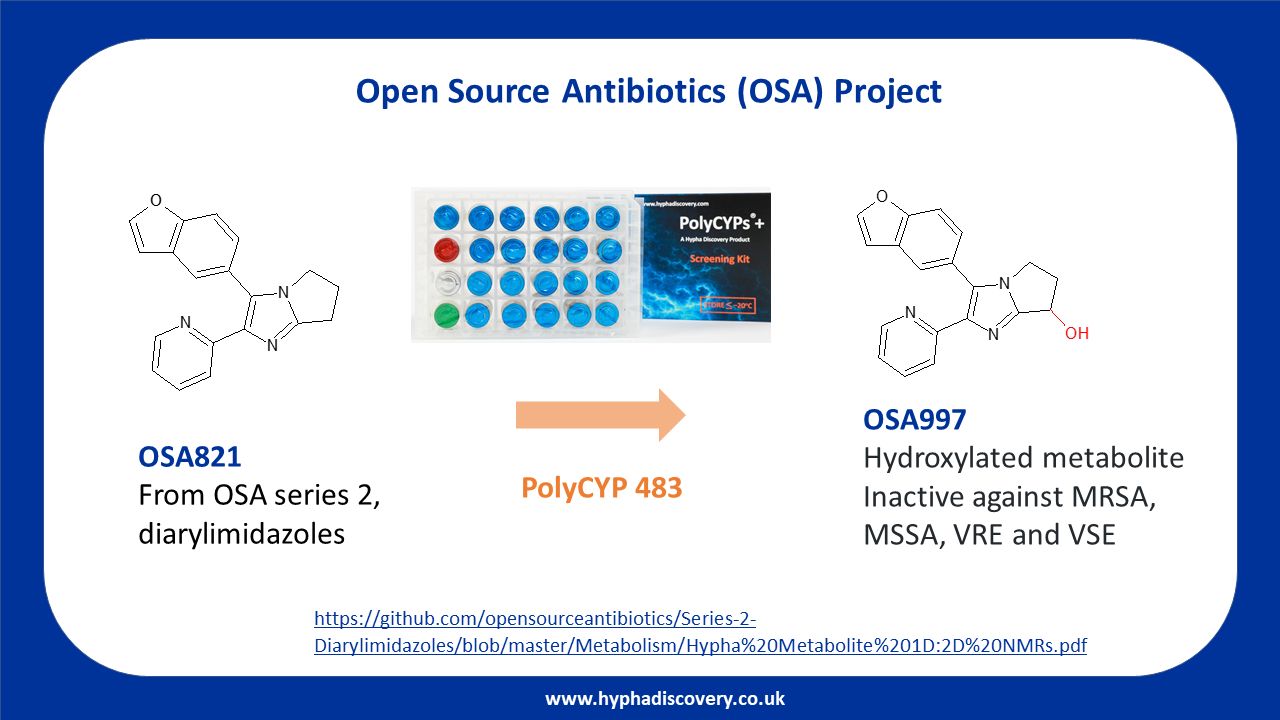
I'm mentioning this because it's just happened again. Some very excellent people at Hypha noticed that we wanted to identify a metabolite of one of the lead molecules in Open Source Antibiotics Series 2. We sent them the molecule with a very light touch MTA (it's all open, after all) and they did their wizardry (that we could not do) and sent us back a sample we were able to purify to prove the location of the oxidation. Fast, efficient, mutually beneficial collaboration, in the open. It was a pleasure working with them.
Naturally there needs to be a trusting relationship between company and project, though the openness helps with that. Often the productive relationship is between the people in the company and the project. Because it's open, there are often zero, or light touch, contractual obligations - this is both good (simple) and bad (can't require anyone to do anything). But it just seems to work so well, repeatedly, and works to everyone's advantage. And it emphasises the point that open science is far from being anti-pharma. Open science includes everyone.
Job Openings in the Lab for Antiviral Drug Discovery
As part of a $65M NIH grant towards new direct acting antivirals, seven posts are available in my group at UCL in central London:
Four synthetic chemistry postdocs https://www.jobs.ac.uk/job/CXH065
One protein science postdoc https://www.jobs.ac.uk/job/CXH076
One Project Manager (not lab based, would particularly suit medicinal chemist with project management experience from industry) https://www.jobs.ac.uk/job/CXH100
One Laboratory Manager https://www.jobs.ac.uk/job/CXH049
The team, working closely with colleagues at the University of North Carolina, Chapel Hill, constitutes the Medicinal Chemistry Core of the READDI-AViDD consortium that aims to improve our preparedness through the design and validation of small molecule binders of proteins from viruses of pandemic potential. Open science governs both the Medchem Core and the Discovery Core (headed by Cheryl Arrowsmith in Toronto), and we will work with our extended partners (including Janssen and Takeda) to translate these molecules into inexpensive therapeutics.
Funding is secured to April 2025, with the possibility of extension for a further two years.
Closing date for all applications is Feb 28th. Start dates: ASAP.
Informal queries can be sent to me at [email protected].
===
Just on a personal note, this is a really exciting initiative:
- It's important, interesting, enabling research, since we're going after novel and under-studied proteins.
- It's a great team, involving Tim Willson and Ralph Baric as consortium leads at UNC, but a host of other excellent researchers at UNC and beyond.
- It's all open science, meaning everything the medchem and discovery cores touch is public domain.
We've already gotten started with a SARS-CoV-2 Nsp13 project over at https://github.com/StructuralGenomicsConsortium/CNP4-Nsp13-C-terminus-B and are at the exciting stage of having novel hits vs protein to explore. The above roles will allow a huge expansion of the effort, against diverse other targets. We're particularly well supported at UCL to do this kind of research, and in my dept at the School of Pharmacy we invested, in 2022, £2M in new high throughput NMR and MS systems precisely to enable faster analysis of protein-small molecule interactions. In 2023 we obtained a new Creoptix Wave, to, which will be very useful throughout the project.
So come and join the team! The synthetic chemistry postdocs will drive the day-to-day (compound synthesis, design and puchase), the protein science postdoc will work on establishing and running suitable biophysical and enzymatic assays, the lab manager will keep everything ship shape and the project manager will oversee all activities in concert with the Chapel Hill team. There's money for regular travel to meet with the rest of the consortium. Full JDs are in the above links.
And you'd be working alongside the existing group - this lovely collection of talented people:
The Sir James Murray Student Champions
Open science projects need leadership to move things forward.
The leadership can be scientific - someone who is the main coordinator, perhaps a founder, or the originator fo the original data. But an essential aspect of leadership is project management. Someone to ensure that tasks are clearly assigned. That meetings happen. That the project can be easily understood by newcomers.
In thinking about scaling up open science projects towards something ambitious like Target 2035, I was reflecting on this requirement for effective management in each project. In my experience the senior academics involved in such open projects are often so distracted with other aspects of their job that they don't make time to take care of the important, more public, aspects of the open science. Similarly, the lab students involved in the open projects can be so caught up in doing experiments, writing them up, interpreting data etc that they don't have time for the community aspects of the work, such as communicating project needs, or chasing others for progress on agreed actions.
The ideal solution is an enthusiastic student, perhaps working on a different project, or writing up a thesis, or someone who was recently involved in research but who is now doing something else - one can think of all manner of scenarios. But someone willing to commit some time to managing the project. A kind of scientific cat-herder who can help keep people focussed, who can act as a collector of community inputs, a verifier of incoming data or an advertiser of current project requirements. Someone able to help with writing manuscripts, help with maintaining the project's Github presence. Someone who would benefit from a structured interaction with a wider research team and whose career and training might benefit from the experience. I've worked with many students in open projects who are really good at this and it makes a huge difference.
I wanted to convey the importance of the role - to give it a name that reflected that importance.
To myself I started calling it the Pied Piper role. A charismatic coordinator, leading others along a road to somewhere fun. But then I checked the story, and the Pied Piper leads children to their deaths. No good at all.
A better role model is Sir James Murray, the visionary curator of public contributions in the creation of the Oxford English Dictionary (OED).
Attribution: By Unknown author. - https://www.amazon.com/Oxford-English-Dictionary-Vol-boxes/dp/0195219422, Public Domain, https://commons.wikimedia.org/w/index.php?curid=6715523
This picture of Sir James from Wikipedia captures things beautifully - he is surrounded by thousands of letters from people helping to build the dictionary by suggesting the first uses of words. The story is well told in the book The Professor and the Madman, and now in a movie with Mel Gibson starring as the man himself. The story was made famous by virtue of one of the most prolific contributors, William Minor, who was an inmate in an insane asylum and it is this aspect - that the OED's crowdsourcing exercise is contributor-blind - that stuck in my mind when I read about it many years ago. Open science projects often receive inputs from all quarters, and often involve conversations between senior researchers and junior students. What matters is not the identity of the contributor but the quality of the work. Just as for the construction of the OED.
So I wanted to call these open science student project managers Sir James Murray Student Champions.
Now, one cannot simply take someone's name without permission. I sought, and obtained, approval from two senior members of the Murray family who were both interested in the project and gracious with their time. Oswyn Murray is a retired Classics don from Oxford (and Prime Minister Boris Johnson's former tutor) and Jim Murray is Professor of Biosciences at Cardiff University. I'm grateful to them for their consideration of my request and for their time in allowing me to explain the idea. An important part of their approval was that all results of open science projects go into the public domain as a public good, for the benefit of everyone. And that the project is contributor-blind. Sir James was, as I understand it, particularly pleased by the involvement of female contributors to the OED and seeing the diversity of the contributors in open science projects is one of the most gratifying aspects of removing barriers to worldwide participation.
So we are inviting students to apply for the role of the Sir James Murray Student Champion for Chemistry Networks Projects. To ensure some continuity we're suggesting the role is one year long in the first instance, with the possibility of renewal if things go well. I hope (and kind of expect) that the Champions will form a squad, sharing expertise and ideas, and that when we're done with this virus, that the squad can get together.
Do I think this will work? Yes, I do. I'm an optimistic realist about open science - what works, what does not. An emblem of that optimism derives from what happened out the front of Sir James' house in Oxford. He received so many contributions from people, and he sent out so many letters, that the Post Office installed a pillarbox right outside his house. It's still there.
Attribution: Wikipedia, ceridwen / Sir James Murray's letterbox, CC BY-SA 2.0
If you've comments on this idea, please write below. If you're interested in actually doing the role, send me a mail, or email [email protected] or check out the relevant SGC page and there is more info on the various tasks involved. I think we're going to build something really extraordinary together. (Update: Chemistry World have featured the idea.)
Professor Chris Abell
My PhD supervisor, Professor Chris Abell, passed away, too soon, last year. The awful news brought back a flood of memories of my time with him, which in turn made me try to figure out why I felt a rush of gratitude. Gratitude that it is now too late to express to him. I am a PhD supervisor. I often wonder about the nature of the impact I have had, or not, on my students as they leave the lab and go out into the wider world. Such departures between mentor and student are hugely emotional, and I probably ought to show it much more than I do. Sometimes students express their feelings directly, years later. This always means a huge amount to me. I spent a fun evening with Chris and Katherine in Madrid in 2015 and I wish I'd taken the opportunity to say Thank You. I think about all this particularly keenly as my own boys are growing up. The creative, slow-burn impact one might have on how people grow.
It's difficult to frame coherent thoughts on the mentor-student relationship, and complicated to write them down, but chemists are supposed to be good at distillation, so:
-
Chris was generous in spirit. He wanted the best for his students. He would give, intellectually, without thought of reward.
-
He loved ideas. And he loved talking about ideas. He just loved that part of life. He would frequently accompany his research group to the tea room (ah, the old tea room) during breaks and engage with whatever nonsense we were talking about. I remember once describing some theory I had about aluminium metal causing Alzheimer's, and lightheartedly suggested I would not be scrubbing my pans too hard any more. Chris listened and kind of nodded, like he thought this was plausible, which at the time really freaked me out.
This picture is how I remember Chris most fondly - sitting somewhere, fully engaged in conversation about something new and interesting.
I remember one beautiful summer evening when we were invited for food and drinks at his house. I recovered from recounting to Katherine some particularly inappropriate Seinfeld moment and followed people into the garden, where we sat on the grass listening to Chris explain what he knew of the shikimate pathway and why Roundup worked. The sun set, the blue sky deepened and the breeze rustled the flowers as he sketched pathways on an easel. He held this event for no reason other than it was fun to talk about interesting things. It remains for me one of those "this is what academia is meant to be" moments.
-
He was supportive. He would often play Devil's Advocate, forcefully. It was important for students to withstand this, and to realise that such sparring was the most important form of intellectual support. During a PhD we screw up, or we coast and lose focus. We apply for jobs and we make mistakes. Chris was unfailingly supportive and had a good word to say no matter what. I'd love to list a selection of examples, but these will probably all remain private moments between me and him, despite their significance to me and the insights they give to who he was.
-
He was laissez-faire. So was my postdoc supervisor Paul Bartlett, actually. In turn it's made me fairly hands off. Some people don't thrive in such an environment, but I saw how this can make people develop their own ideas, enthusiasm and scientific direction. Chris was around if you needed him - he'd sometimes come into the lab and just sit down without a reason. There was no particular need to talk about recent results, and I think sometimes the details could bore him. This led his students to talk to each other to solve technical and scientific problems quickly. It created a thriving intellectual atmosphere where we had plenty of time to figure out what we were interested in. The challenge was whether we could take an idea to Chris that was more interesting than the thing he had in his head, that day. Hours sitting in recliners in the Chemistry library reading goodness knows what, and I knew he'd approve.
-
He laughed. Loud. Jeez, I miss that.
I know it's too late now, but thank you, Chris.
Donating Molecules to Open Source Drug Discovery Projects
Open drug discovery projects have several advantages, and one of those needs to be the ease with which people can contribute physical samples for evaluation: either molecules they have made for the project specifically, or molecules that may be useful and are just sitting in a freezer.

(Image adapted from Nick Smith photography, CC BY-SA 3.0, via Wikimedia Commons)
People have donated molecules to open projects before in various ways, for example synthesizing them in their own labs and having them tested nearby (e.g., Patrick Thomson and Chase Smith over in OSM OpenSourceMalaria/OSM_To_Do_List#487), or digging them out of the freezer and sending them in (e.g., Helen Hailes for OSM Series 5, and DNDi (opensourceantibiotics/Series-2-Diarylimidazoles#27) and Lori Ferrins (opensourceantibiotics/Series-2-Diarylimidazoles#48) in OSA Series 2). Lots of researchers are interested in seeing whether their fridge contains something useful for drug discovery, serendipitously or because of perceived molecular similarity. But I think lots of those people worry about getting bogged down in legal paperwork, and some industry folks are maybe worried that if they donate molecules they are somehow obligated to do more.
We've never had any clear arrangements for these sample donations, but as we move to scale up more open projects, clarity is needed. The general guidelines for open projects are as described in the Six Laws, and the overall license is always CC-BY-4.0 (the Wikipedia licence) but these don't adequately cover physical samples, so we need something fresh. We'd like to avoid, if possible, the need for MTAs (even openMTA, though I'm open to be persuaded just to adopt this) just because that slows things down and increases transactional costs dramatically. There are some good rules available for requesting compounds that have been devised by the SGC (Paper), i.e. where someone wants to receive a chemical probe from the SGC (the reverse of what we're talking about here). The nice thing about the SGC terms is that the user has to agree to the rules as part of the request. Similarly for open drug discovery projects: the rules need to be agreed to as part of the submission.
We got chatting about this in a recent Open Source Antibiotics meeting opensourceantibiotics/Series-2-Diarylimidazoles#45 and have consulted on the guidelines.
The most recent version is here.
The guidelines cover things like the obligations on the contributor and the open project, potential future uses of the samples, and what happens if future data are derived.
These rules will be updated onto the current molecule submission page for OSA, and they can be cloned for other open projects.
I'd love to hear any suggestions (particularly from potential industry contributors) please comment below or tweet us at or get in touch with me by email or on LinkedIn.
Do these rules work, and is there anything that's missing?
(there are lots of ways in which we might improve the process of connecting molecules with the people who might need them, ranging from use of commercial vendors through to Molecular Craigslist type sites that I've been talking about forever through to newer things I and others are thinking about, but whatever we do we'll need simple submission guidelines, and terms).
Open Source Antibiotics Paper Number 1
Open Source Antibiotics (OSA) began in November 2018 with a coffee in the Green Building at the Old Road site at the University of Oxford. I was talking to Paul Brennan and Frank von Delft about taking fragment hits to lead compounds - there were few good examples of this back then - and we thought we should get something going. I suggested we open things up to the community. That project became the first project of OSA and is focussed on the mur ligases. Though we've very cool recent data on that project, we never (despite the best efforts of my talented postdoc at the time, Fahima Idris) got the fragments to go anywhere, but that's a different (and ongoing) story.
Though we were busy making the economic/policy argument about the value of open source methods for new antibiotic development (essentially, there is no market, so let's open it up) I was thinking we should start a second OSA series. One of the great potential benefits of a really open approach is its ability to immortalise teenage projects: work that has gone pretty far, and has obvious promise, but that is not yet fully mature. Let's say you have a series you're working on that looks OK but you either can't raise money or you can't quite get the potency high enough, or you have other things that are more pressing. You can't publish the work, and you think that you need to head off in a different direction. Do you just dispose of the data and forget all about it? Why not put the data (and your ideas) somewhere that other people can work on it all? Maybe someone out there has an idea, or can raise some funds, to take things to the next step. Immortalisation.
Just such a thing happened to OSA, and Series 2 was born. 3 years later, it's published.
The project started with a mail from Bill Zuercher, then at UNC Chapel Hill. He and the team there had evaluated a bunch of molecules they'd found in another project against bacteria, using CO-ADD as their screening resource. The data looked interesting and worth exploring. I managed to get a grant from something called Pharmalliance, which is a coalition of the pharmacy schools from UCL, UNC and Monash. We worked for a while doing some resynthesis and basic analog synthesis towards some kind of SAR.
The notable thing about this project was the feature we've often seen in projects of this kind, whereby a funded core in an open science project can attract contributors, in this case really valuable expertise from academia (Lori Ferrins from Northeastern as an excellent example), Industry (Hypha Discovery who worked with us on metabolite ID), NGOs (thank you, DNDi) consultancy (the ever-wise Chris Swain) and the public (citizen scientist Anthony Sama).
The regular meetings over the course of the pandemic were a real pleasure to run and record and helped keep everyone connected and on the same page.

These are interesting compounds which still have promise. Though we've stopped the project (in part, as always, through not having further funding) the project remains ready to re-activate. If you're a PhD or Masters student wanting something cool to work on, you could extend what we've done in full knowledge that you're in possession of all the facts and would not be unintentionally duplicating something we did that did not go anywhere. If you wanted to do the next key experiments, some of which we list at the end of the paper, you'd be more than welcome (the licence is CC-BY-4.0, so you don't need to ask permission - just cite your source) and you could use the data we've generated as the basis for your application for more resources. On a personal note I was frustrated that we never managed to figure out the bacterial protein target. I'd love to know what that is!
Recommend Projects
-
 React
React
A declarative, efficient, and flexible JavaScript library for building user interfaces.
-
Vue.js
🖖 Vue.js is a progressive, incrementally-adoptable JavaScript framework for building UI on the web.
-
 Typescript
Typescript
TypeScript is a superset of JavaScript that compiles to clean JavaScript output.
-
TensorFlow
An Open Source Machine Learning Framework for Everyone
-
Django
The Web framework for perfectionists with deadlines.
-
Laravel
A PHP framework for web artisans
-
D3
Bring data to life with SVG, Canvas and HTML. 📊📈🎉
-
Recommend Topics
-
javascript
JavaScript (JS) is a lightweight interpreted programming language with first-class functions.
-
web
Some thing interesting about web. New door for the world.
-
server
A server is a program made to process requests and deliver data to clients.
-
Machine learning
Machine learning is a way of modeling and interpreting data that allows a piece of software to respond intelligently.
-
Visualization
Some thing interesting about visualization, use data art
-
Game
Some thing interesting about game, make everyone happy.
Recommend Org
-
Facebook
We are working to build community through open source technology. NB: members must have two-factor auth.
-
Microsoft
Open source projects and samples from Microsoft.
-
Google
Google ❤️ Open Source for everyone.
-
Alibaba
Alibaba Open Source for everyone
-
D3
Data-Driven Documents codes.
-
Tencent
China tencent open source team.