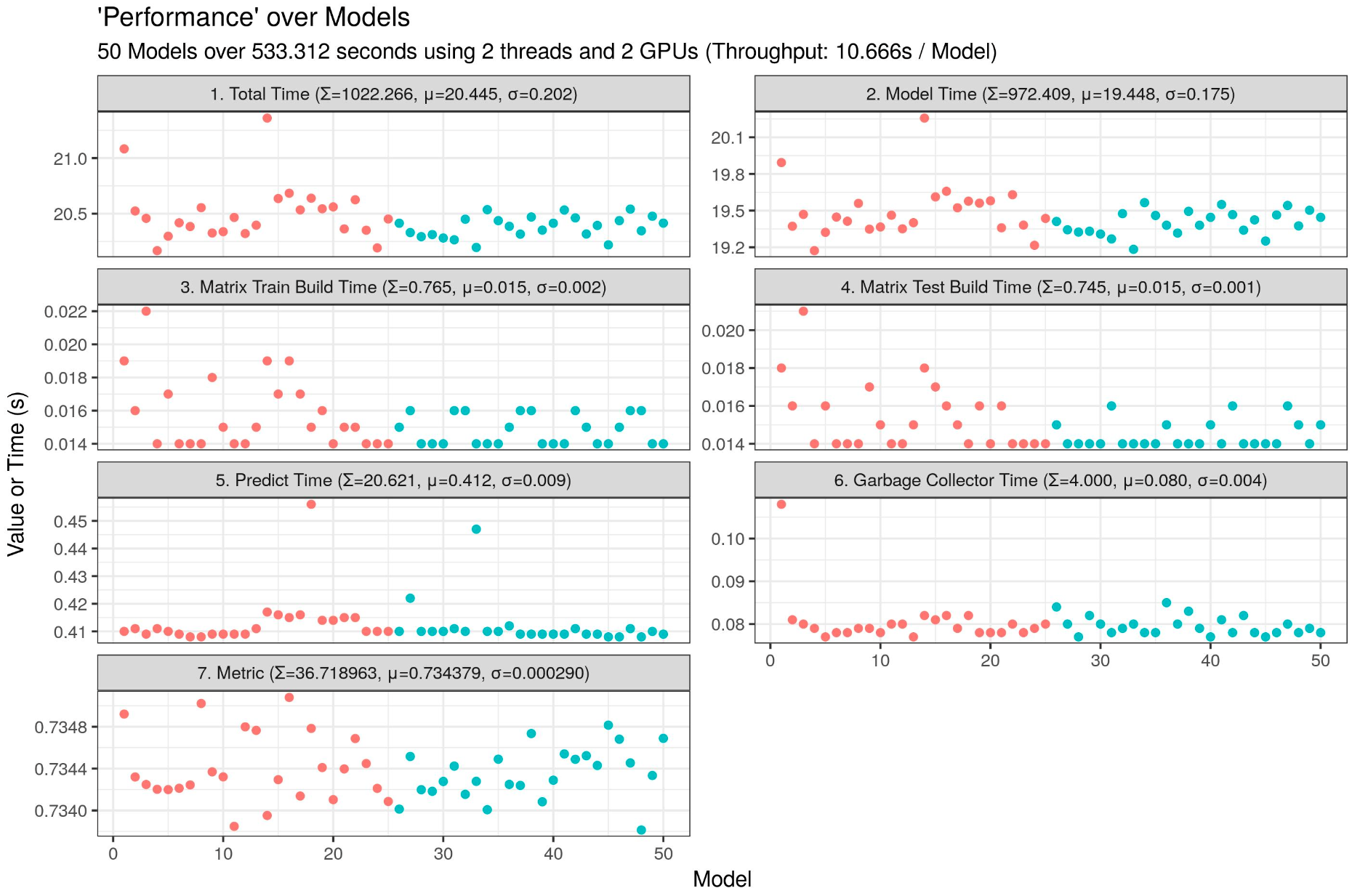
I am currently testing a LightGBM script which can run 40 LightGBM on one 4GB GPU (only 2690 MB used).
Throughput: 1.330s / model, insane speed! Imagine on multiple GPUs...
You need 67.25 MB per thread for LightGBM on airline 0.1m data. It's about 10x lower than xgboost. Maybe @RAMitchell can get some ideas to optimize xgboost for very sparse data on GPU, as it could be a common case (xgboost does not support categoricals). LightGBM handles 16 bins, 64 bins, and 256 bins as shown here: https://github.com/microsoft/LightGBM/tree/master/src/treelearner/ocl
# Sets OpenMP to 1 thread by default
Sys.setenv(OMP_NUM_THREADS = 1)
suppressMessages({
library(optparse)
library(data.table)
library(parallel)
library(lightgbm)
library(Matrix)
})
args_list <- list(
optparse::make_option("--parallel_threads", type = "integer", default = 1, metavar = "Parallel CPU Threads",
help="Number of threads for parallel training for CPU (automatically changed if using GPU), should be greater than or equal to parallel_gpus * gpus_threads [default: %default]"),
optparse::make_option("--model_threads", type = "integer", default = 1, metavar = "Model CPU Threads",
help = "Number of threads for training a single model, total number of threads is parallel_threads * model_threads [default: %default]"),
optparse::make_option("--parallel_gpus", type = "integer", default = 0, metavar = "Parallel GPU Threads",
help = "Number of GPUs to use for parallel training, use 0 for no GPU [default: %default]"),
optparse::make_option("--gpus_threads", type = "integer", default = 0, metavar = "Model GPU Threads",
help = "Number of parallel models to train per GPU (uses linearly more RAM), use 0 for no GPU [default: %default]"),
optparse::make_option("--number_of_models", type = "integer", default = 1, metavar = "Number of Models",
help = "Number of models to train in total [default: %default]"),
optparse::make_option("--wkdir", type = "character", default = "", metavar = "Working Directory",
help = "The working directory, do NOT forget it! [default: \"%default\"]"),
optparse::make_option("--train_file", type = "character", default = "", metavar = "Training File",
help = "The training file to use relative to the working directory (or an absolute path), do NOT forget it! [default: \"%default\"]"),
optparse::make_option("--test_file", type = "character", default = "", metavar = "Testing file",
help = "The testing file to use relative to the working directory (or an absolute path), do NOT forget it! [default: \"%default\"]"),
optparse::make_option("--output_dir", type = "character", default = "", metavar = "Output Directory",
help = "The output directory for files (or an absolute path), do NOT forget it! [default: \"%default\"]"),
optparse::make_option("--output_csv", type = "logical", default = TRUE, metavar = "Output CSV File",
help = "Outputs results as a CSV file [default: %default]"),
optparse::make_option("--output_chart", type = "character", default = "jpeg", metavar = "Plot File Format",
help = "Outputs results as a chart using the desired format, can be any of: \"none\" (for no chart), \"eps\", \"ps\", \"tex\" (pictex), \"pdf\", \"jpeg\", \"tiff\", \"png\", \"bmp\", \"svg\", \"wmf\" (Windows only) [default: \"%default\"]"),
optparse::make_option("--args", type = "logical", default = FALSE, metavar = "Argument Check",
help = "Prints the arguments passed to the R script and exits immediately [default: %default]")
)
# Force data.table as 1 thread in case you are using Fork instead of Sockets (gcc: fork X in process Y when process Y used OpenMP once, fork X cannot use OpenMP otherwise it hangs forever)
data.table::setDTthreads(1)
if (interactive()) {
# Put some parameters if you wish to test once...
my_gpus <- 1L
my_gpus_threads <- 40L
my_threads <- 1L # parallel::detectCores() - 1L
my_threads_in_threads <- 1L
my_runs <- 40L
my_train <- "train-0.1m.csv"
my_test <- "test.csv"
my_output <- "./output"
my_csv <- TRUE
my_chart <- "jpeg"
# my_cpu <- system("lscpu | sed -nr '/Model name/ s/.*:\\s*(.*) @ .*/\\1/p' | sed ':a;s/ / /;ta'")
# CHANGE: 0.1M = GPU about 958 MB at peak... choose wisely (here, we are putting 4 models per GPU)
if (my_gpus > 0L) {
# my_threads <- min(my_gpus * my_gpus_threads, my_threads)
my_threads <- my_gpus * my_gpus_threads
}
} else {
# Old school method... obsolete
# DIR="$( cd "$( dirname "${BASH_SOURCE[0]}" )" && pwd )"
# Rscript bench_file.R 1 1 0 0 25 ${DIR} ../train-0.1m.csv ../test.csv
# args <- commandArgs(trailingOnly = TRUE)
#
# setwd(args[6])
# my_gpus <- args[3]
# my_gpus_threads <- args[4]
# my_threads <- args[1]
# my_threads_in_threads <- args[2]
# my_runs <- args[5]
# my_train <- args[7]
# my_test <- args[8]
# DIR="$( cd "$( dirname "${BASH_SOURCE[0]}" )" && pwd )"
# Rscript bench_lgb_test.R --parallel_threads=1 --model_threads=1 --parallel_gpus=0 --gpus_threads=0 --number_of_models=25 --wkdir=${DIR} --train_file=../train-0.1m.csv --test_file=../test.csv --output_dir=./output --output_csv=TRUE --output_chart=jpeg --args=TRUE
# Rscript bench_lgb_test.R --parallel_threads=1 --model_threads=1 --parallel_gpus=0 --gpus_threads=0 --number_of_models=25 --wkdir=${DIR} --train_file=../train-0.1m.csv --test_file=../test.csv --output_dir=./output --output_csv=TRUE --output_chart=jpeg
args <- optparse::parse_args(optparse::OptionParser(option_list = args_list))
setwd(args$wkdir)
my_gpus <- args$parallel_gpus
my_gpus_threads <- args$gpus_threads
my_threads <- args$parallel_threads
my_threads_in_threads <- args$model_threads
my_runs <- args$number_of_models
my_train <- args$train_file
my_test <- args$test_file
my_output <- args$output_dir
my_csv <- args$output_csv
my_chart <- args$output_chart
if (my_gpus > 0L) {
# my_threads <- min(my_gpus * my_gpus_threads, my_threads)
my_threads <- my_gpus * my_gpus_threads
args$parallel_threads <- my_threads
}
if (args$args) {
print(args)
stop("\rArgument check done.")
}
}
# Load data and do preprocessing
cat("[", format(Sys.time(), "%a %b %d %Y %X"), "] [Data] Loading data.\n", sep = "")
d_train <- fread(my_train, showProgress = FALSE)
d_test <- fread(my_test, showProgress = FALSE)
invisible(gc(verbose = FALSE))
cat("[", format(Sys.time(), "%a %b %d %Y %X"), "] [Data] Transforming data.\n", sep = "")
X_train_test <- sparse.model.matrix(dep_delayed_15min ~ . -1, data = rbindlist(list(d_train, d_test))); invisible(gc(verbose = FALSE))
n1 <- nrow(d_train)
n2 <- nrow(d_test)
X_train <- X_train_test[1L:n1,]; invisible(gc(verbose = FALSE))
X_test <- X_train_test[(n1 + 1L):(n1 + n2),]; invisible(gc(verbose = FALSE))
labels_train <- as.numeric(d_train$dep_delayed_15min == "Y")
labels_test <- as.numeric(d_test$dep_delayed_15min == "Y")
rm(d_train, d_test, X_train_test, n1, n2); invisible(gc(verbose = FALSE))
# CHANGE: metric function
metric <- function(preds, labels) {
x1 <- as.numeric(preds[labels == 1])
n1 <- as.numeric(length(x1))
x2 <- as.numeric(preds[labels == 0])
n2 <- as.numeric(length(x2))
r <- rank(c(x1,x2))
return((sum(r[1:n1]) - n1 * (n1 + 1) / 2) / (n1 * n2))
}
# CHANGE: trainer function
trainer <- function(x, row_sampling, col_sampling, max_depth, n_iter, learning_rate, nbins, nthread, n_gpus, gpu_choice, objective) {
if (n_gpus > 0) {
matrix_train_time <- system.time({
dlgb_train <- lgb.Dataset(data = X_train, label = labels_train, nthread = nthread, device = "gpu")
lgb.Dataset.construct(dlgb_train)
})[[3]]
matrix_test_time <- system.time({
dlgb_test <- lgb.Dataset(data = X_test, label = labels_test, nthread = nthread, device = "gpu") # Completely useless in practice
lgb.Dataset.construct(dlgb_test) # Completely useless in practice
})[[3]]
} else {
matrix_train_time <- system.time({
dlgb_train <- lgb.Dataset(data = X_train, label = labels_train, nthread = nthread)
lgb.Dataset.construct(dlgb_train)
})[[3]]
matrix_test_time <- system.time({
dlgb_test <- lgb.Dataset(data = X_test, label = labels_test, nthread = nthread) # Completely useless in practice
lgb.Dataset.construct(dlgb_test) # Completely useless in practice
})[[3]]
}
if (n_gpus > 0) {
model_time <- system.time({
set.seed(x) # Useless
model_train <- lightgbm::lgb.train(data = dlgb_train,
objective = objective,
nrounds = n_iter,
num_leaves = (2 ^ max_depth) - 1,
max_depth = max_depth,
learning_rate = learning_rate,
bagging_fraction = row_sampling,
bagging_seed = x,
bagging_freq = 1,
feature_fraction = col_sampling,
feature_fraction_seed = x,
num_threads = nthread,
device = "gpu",
gpu_platform_id = 0,
gpu_device_id = gpu_choice,
max_bin = nbins,
verbose = -1)
})[[3]]
} else {
model_time <- system.time({
set.seed(x)
model_train <- lightgbm::lgb.train(data = dlgb_train,
objective = objective,
nrounds = n_iter,
num_leaves = (2 ^ max_depth) - 1,
max_depth = max_depth,
learning_rate = learning_rate,
bagging_fraction = row_sampling,
bagging_seed = x,
bagging_freq = 1,
feature_fraction = col_sampling,
feature_fraction_seed = x,
num_threads = nthread,
device = "cpu",
max_bin = nbins,
verbose = -1)
})[[3]]
}
pred_time <- system.time({
model_predictions <- predict(model_train, data = X_test)
})[[3]]
perf <- metric(preds = model_predictions, labels = labels_test)
rm(model_train, model_predictions, dlgb_train, dlgb_test)
gc_time <- system.time({
invisible(gc(verbose = FALSE))
})[[3]]
return(list(matrix_train_time = matrix_train_time, matrix_test_time = matrix_test_time, model_time = model_time, pred_time = pred_time, gc_time = gc_time, perf = perf))
}
# Parallel Section
cat("[", format(Sys.time(), "%a %b %d %Y %X"), "]", " [Parallel] ", my_threads, " Process(es) Creation Time: ", sprintf("%04.03f", system.time({cl <- makeCluster(my_threads)})[[3]]), "s\n", sep = "")
cat("[", format(Sys.time(), "%a %b %d %Y %X"), "]", " [Parallel] Sending Hardware Specifications Time: ", sprintf("%04.03f", system.time({clusterExport(cl = cl, c("my_threads", "my_gpus", "my_threads_in_threads"))})[[3]]), "s\n", sep = "")
invisible(parallel::parLapply(cl = cl, X = seq_len(my_threads), function(x) {
Sys.sleep(time = my_threads / 20) # Prevent file clash on many core systems (typically 50+ threads might attempt to read exactly at the same time the same file, especially if the disk is slow)
suppressPackageStartupMessages(library(lightgbm))
suppressPackageStartupMessages(library(Matrix))
suppressPackageStartupMessages(library(data.table))
id <<- x
}))
cat("[", format(Sys.time(), "%a %b %d %Y %X"), "]", " [Parallel] Sending Data Time: ", sprintf("%04.03f", system.time({clusterExport(cl = cl, c("trainer", "metric", "X_train", "X_test", "labels_train", "labels_test", "my_threads"))})[[3]]), "s\n", sep = "")
# Having issues? In a CLI: sudo pkill R
time_finish <- system.time({
time_all <- parallel::parLapplyLB(cl = cl, X = seq_len(my_runs), function(x) {
if (my_gpus == 0L) {
gpus_to_use <- 0
gpus_allowed <- 0
} else {
gpus_to_use <- (id - 1) %% my_gpus
gpus_allowed <- 1
}
speed_out <- system.time({
speed_in <- trainer(x = x,
row_sampling = 0.9,
col_sampling = 0.9,
max_depth = 6,
n_iter = 500,
learning_rate = 0.05,
nbins = 255,
nthread = my_threads_in_threads,
n_gpus = gpus_allowed,
gpu_choice = gpus_to_use,
objective = "binary")
})[[3]]
rm(gpus_to_use)
return(list(total = speed_out, matrix_train_time = speed_in$matrix_train_time, matrix_test_time = speed_in$matrix_test_time, model_time = speed_in$model_time, pred_time = speed_in$pred_time, gc_time = speed_in$gc_time, perf = speed_in$perf))
})
})[[3]]
# Clearup all R sessions from this process, except the master
stopCluster(cl)
closeAllConnections()
rm(cl, metric, trainer, X_train, X_test, labels_train, labels_test); invisible(gc(verbose = FALSE))
cat("[", format(Sys.time(), "%a %b %d %Y %X"), "]", " [Parallel] Total Time: ", sprintf("%04.03f", time_finish), "s\n", sep = "")
# Gather Data
# Get data
time_total <- unlist(lapply(time_all, function(x) {round(x[[1]], digits = 3)}))
matrix_train_time <- unlist(lapply(time_all, function(x) {round(x[[2]], digits = 3)}))
matrix_test_time <- unlist(lapply(time_all, function(x) {round(x[[3]], digits = 3)}))
model_time <- unlist(lapply(time_all, function(x) {round(x[[4]], digits = 3)}))
pred_time <- unlist(lapply(time_all, function(x) {round(x[[5]], digits = 3)}))
gc_time <- unlist(lapply(time_all, function(x) {round(x[[6]], digits = 3)}))
perf <- unlist(lapply(time_all, function(x) {round(x[[7]], digits = 6)}))
# Put all data together
time_table <- data.table(Run = seq_len(my_runs),
time_total = time_total,
matrix_train_time = matrix_train_time,
matrix_test_time = matrix_test_time,
model_time = model_time,
pred_time = pred_time,
gc_time = gc_time,
perf = perf)
if (my_csv) {
fwrite(time_table, paste0(my_output, "/ml-perf_lgb_gbdt_", substr(my_train, 1, nchar(my_train) - 4), "_", my_threads, "Tx", my_threads_in_threads, "T_", my_gpus, "GPU_", my_runs, "m_", sprintf("%04.03f", time_finish), "s.csv"))
}
# Analyze Data
if (my_chart != "none") {
suppressMessages({
library(ggplot2)
library(ClusterR)
})
# Create time series matrix
time_table_matrix <- apply(as.matrix(time_table[, 2:8, with = FALSE]), MARGIN = 2, function(x) {
y <- cumsum(x)
y / max(y)
})
# Compute optimal number of non-parametric clusters
clusters <- Optimal_Clusters_Medoids(data = time_table_matrix,
max_clusters = 2:10,
distance_metric = "manhattan",
criterion = "silhouette",
threads = 1,
swap_phase = TRUE,
verbose = FALSE,
plot_clusters = FALSE,
seed = 1)
# Compute clusters
clusters_selected <- Cluster_Medoids(data = time_table_matrix,
clusters = 1 + which.max(unlist(lapply(clusters, function(x) {x[[3]]}))),
distance_metric = "manhattan",
threads = 1,
swap_phase = TRUE,
verbose = FALSE,
seed = 1)
time_table[, Cluster := as.character(clusters_selected$clusters)]
# Melt data
time_table_vertical <- melt(time_table, id.vars = c("Run", "Cluster"), measure.vars = c("time_total", "matrix_train_time", "matrix_test_time", "model_time", "pred_time", "gc_time", "perf"), variable.name = "Variable", value.name = "Value", variable.factor = FALSE, value.factor = FALSE)
# Rename melted variables to have details in chart
time_table_vertical[Variable == "time_total", Variable := paste0("1. Total Time (Σ=", sprintf("%04.03f", sum(Value)), ", μ=", sprintf("%04.03f", mean(Value)), ", σ=", sprintf("%04.03f", sd(Value)), ")")]
time_table_vertical[Variable == "model_time", Variable := paste0("2. Model Time (Σ=", sprintf("%04.03f", sum(Value)), ", μ=", sprintf("%04.03f", mean(Value)), ", σ=", sprintf("%04.03f", sd(Value)), ")")]
time_table_vertical[Variable == "matrix_train_time", Variable := paste0("3. Matrix Train Build Time (Σ=", sprintf("%04.03f", sum(Value)), ", μ=", sprintf("%04.03f", mean(Value)), ", σ=", sprintf("%04.03f", sd(Value)), ")")]
time_table_vertical[Variable == "matrix_test_time", Variable := paste0("4. Matrix Test Build Time (Σ=", sprintf("%04.03f", sum(Value)), ", μ=", sprintf("%04.03f", mean(Value)), ", σ=", sprintf("%04.03f", sd(Value)), ")")]
time_table_vertical[Variable == "pred_time", Variable := paste0("5. Predict Time (Σ=", sprintf("%04.03f", sum(Value)), ", μ=", sprintf("%04.03f", mean(Value)), ", σ=", sprintf("%04.03f", sd(Value)), ")")]
time_table_vertical[Variable == "gc_time", Variable := paste0("6. Garbage Collector Time (Σ=", sprintf("%04.03f", sum(Value)), ", μ=", sprintf("%04.03f", mean(Value)), ", σ=", sprintf("%04.03f", sd(Value)), ")")]
time_table_vertical[Variable == "perf", Variable := paste0("7. Metric (Σ=", sprintf("%07.06f", sum(Value)), ", μ=", sprintf("%07.06f", mean(Value)), ", σ=", sprintf("%07.06f", sd(Value)), ")")]
cat(sort(unique(time_table_vertical$Variable)), sep = "\n")
# Plot a nice chart
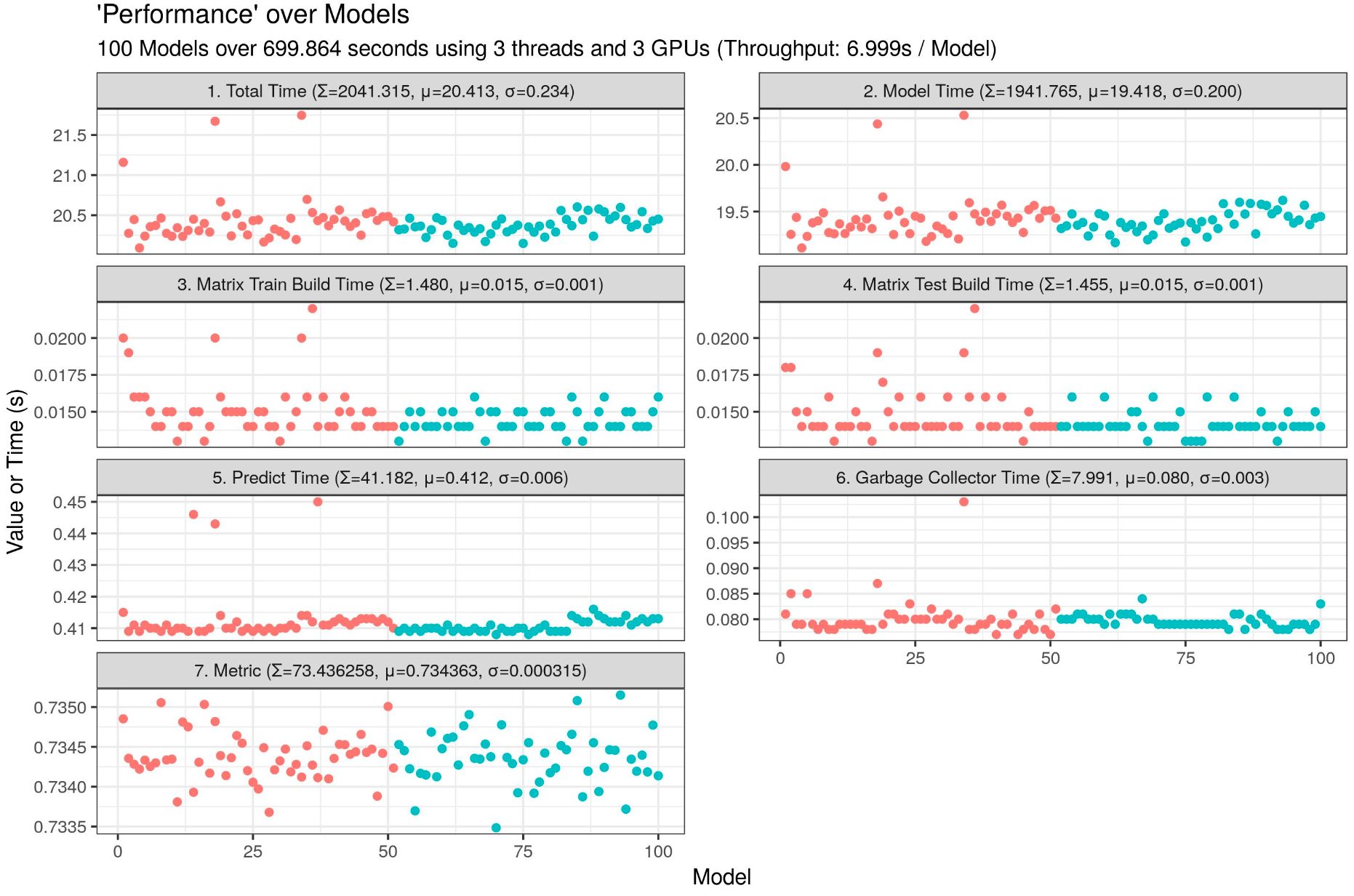
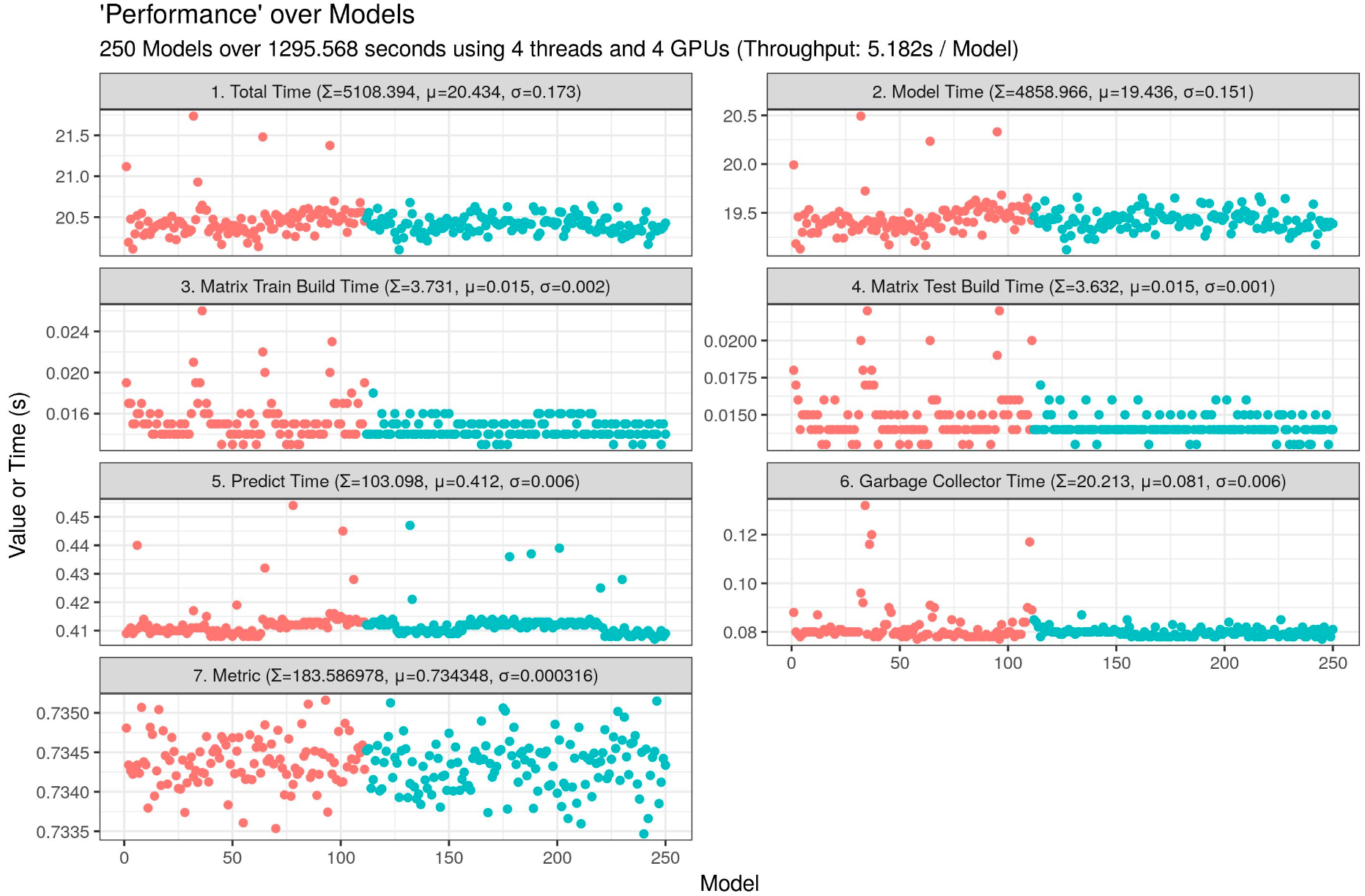
my_plot <- ggplot(data = time_table_vertical, aes(x = Run, y = Value, group = Cluster, color = Cluster)) + geom_point() + facet_wrap(facets = Variable ~ ., nrow = 4, ncol = 2, scales = "free_y") + labs(title = "'Performance' over Models, LightGBM GBDT", subtitle = paste0(my_runs, " Models over ", sprintf("%04.03f", time_finish), " seconds using ", my_threads, " parallel threads, ", my_threads_in_threads, " model threads, and ", my_gpus, " GPUs (Throughput: ", sprintf("%04.03f", time_finish / my_runs), "s / Model", ")"), x = "Model", y = "Value or Time (s)") + theme_bw() + theme(legend.position = "none")
ggsave(filename = paste0(my_output, "/ml-perf_lgb_gbdt_", substr(my_train, 1, nchar(my_train) - 4), "_", my_threads, "Tx", my_threads_in_threads, "T_", my_gpus, "GPU_", my_runs, "m_", sprintf("%04.03f", time_finish), "s.jpg"),
plot = my_plot,
device = my_chart,
width = 24,
height = 16,
units = "cm",
dpi = "print")
if (interactive()) {
print(my_plot)
}
}
cat("[", format(Sys.time(), "%a %b %d %Y %X"), "] Done computations. Quitting R.\n", sep = "")