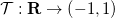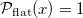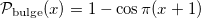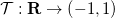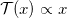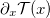Do you usually use sklearn.preprocessing.StandardScaler to normalize your data? Sometimes, the distribution of the data can be so assymetric and heterogeneous that a better transformation is necessary. PrettyScaler acts in the same way as StandardScaler; fits a mapping from raw to normalized distributions. However, PrettyScaler will neatly condense the metric, squeezing all reals into the bound domain (-1,1) and producing a uniform density distribution.
Basically, it will map each 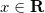
The result of this transformation is a uniform distribution, but PrettyScaler can also output one that generates a bulge-like distribution. Depending on the "kind" argument in the fit() method, the probability density distributions will be:
The mapping constructed by PrettyScaler meets the following requirements:
When to use PrettyScaler:
- The main use case is to preprocess data for neural nets and SVMs
- It is not helpful for CART-based methods (decision trees, random forest, GBM), although it will not hurt either
- It will completely wash out 1-D clustering signal, so don't use it for this purpose
# Make some basic imports
import matplotlib.pyplot as plt;
%matplotlib inline
import numpy as np;
import prettyscaler;
from sklearn.preprocessing import StandardScaler;# let's create some ugly mock data. This will be a mixture
# of a few truncated log-normal distributions:
def create_lognormal_truncated_sample(logmu,logsig,N,sign):
random_sample = np.random.normal(logmu,logsig,size=N)
random_sample = random_sample[sign*random_sample>sign*logmu]
return 10**random_sample
random_sample1 = create_lognormal_truncated_sample(1,1,5000,-1)
random_sample2 = create_lognormal_truncated_sample(5,.3,5000,1)
random_sample3 = create_lognormal_truncated_sample(8,.25,5000,-1)
# Assemble the full data set:
my_data = np.r_[random_sample1,random_sample2, random_sample3]# Take a look at the monster we've created in a log plot
plt.hist(np.log10(my_data[my_data>0]),bins=50);# Break the random sample into train and test sets:
np.random.shuffle(my_data)
test_data, train_data = my_data[::2], my_data[1::2]# Initialize
pts = prettyscaler.PrettyScaler()
# Fit the data. The most important argument you need
# to care about is the wing_percentile. Roughly,
# set it to the fraction of your data that you would
# consider quasi-outliers. For example, wing_percentile=0.01
# will result in fitting the data directly with 98% of the points,
# and a 1% on each side is discarded (the reason for a non-zero
# wing_percentile is that we need a robust computation of the gradient
# at the wing boundary, and there needs to be enough data density).
# The argument sample_size determines the size of the grid
# that will be used to map the distribution densities. As a rule
# of thumb, try to have wing_percentile*sample_size > 10. If sample_size
# happens to be larger that the size of the training data, it will revert
# to the latter.
# Finally, ftol is the kind of the minimum difference enforced between
# consecutive values in the cumulative density array. It is there to make sure
# that the gradient of that array not close to being (numerically) flat anywhere.
#
# This might be a lot to take, but for most cases you can just
# use the default values and not worry about it.
#
# So, let's fit some data:
pts.fit(test_data, wing_percentile=0.01,ftol=1e-5,sample_size=1e3)
# If you want to load these params later, you should save
# them now to file:
pts.save_pickle('fitted_params.pkl')
# To load this state from scratch, you would just have to do:
# pts = prettydata.PrettyData()
# pts.load_pickle('fitted_params.pkl')# Let us plot the input vs output spaces of the function.
# We will highlight the different regions (wings and trunk)
# separately, and examine how well they blend together.
#
# First, gotta create an x-grid for the inputs. For
# better clarity, we'll make it logarithmically spaced:
right_wing_x = 10**np.linspace(np.log10(pts.all_params['rightwing_boundary_value']),np.log10(1e9),100)
left_wing_x = 10**np.linspace(np.log10(1e-4),np.log10(pts.all_params['leftwing_boundary_value']),100)
# This one we can fetch directly from the instance:
trunk_x = pts.all_params['trunk_x']
# Now, compute the outputs
right_wing_y = np.array([pts.transfer_function(u) for u in right_wing_x])
left_wing_y = np.array([pts.transfer_function(u) for u in left_wing_x])
trunk_y = np.array([pts.transfer_function(u) for u in trunk_x])
# Plot the whole thing. The wings are shown in red.
# As mentioned earlier, the trunk (blue part) represents
# an interpolation to the cumulative function of the data.
# The wings are not a direct fit to the data; they are
# asymptotically declining functions that match the value
# and gradient of the trunk at the boundaries.
plt.plot(left_wing_x, left_wing_y, color='r')
plt.plot(right_wing_x, right_wing_y, color='r')
plt.plot(trunk_x, trunk_y, color='b')
plt.ylim(-1.1,1.1)
plt.xscale('log')
plt.yscale('linear')# Zoom into the left wing blend, it's pretty smooth:
plt.plot(left_wing_x, left_wing_y, color='r')
plt.plot(trunk_x, trunk_y, color='b')
plt.xlim(1e-3,5e-1)
plt.ylim(-1,-0.9)
plt.xscale('log')# Zoom into the right wing. Also smooth blend.
plt.plot(right_wing_x, right_wing_y, color='r')
plt.plot(trunk_x, trunk_y, color='b')
plt.xlim(9e7,1.1e8)
plt.ylim(0.95,1)
plt.xscale('linear')# In addition, the gradient should be positive everywhere.
# Assemble full output grid:
test_ygrid = np.r_[left_wing_y, trunk_y, right_wing_y]
# Check that there is no negative gradient:
assert(min(np.gradient(test_ygrid))>=0)
# In the trunk specifically, check that the
# gradient is larger than zero everywhere:
assert(min(np.gradient(trunk_y))>0)# Transform the test data into a flat and bulge spaces:
test_data_flat = pts.transform(test_data, kind='flat')
test_data_bulge = pts.transform(test_data, kind='bulge')Mapping to bulge distribution
# Plot the histogram of the flat representation
plt.hist(test_data_flat, bins=20);# And a histogram of the bulge representation
plt.hist(test_data_bulge, bins=20);