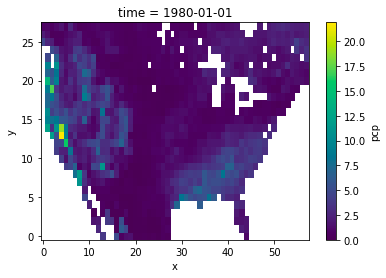
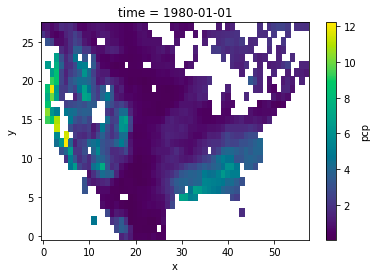
---------------------------------------------------------------------------
TypeError Traceback (most recent call last)
<ipython-input-18-e629ff74bc4d> in <module>()
1 grid = ESMF.Grid(np.array([20000, 15000]),
2 staggerloc=ESMF.StaggerLoc.CENTER,
----> 3 coord_sys=ESMF.CoordSys.SPH_DEG)
~/Research/Computing/miniconda3/envs/sci/lib/python3.6/site-packages/ESMF/util/decorators.py in new_func(*args, **kwargs)
62
63 esmp = esmpymanager.Manager(debug = False)
---> 64 return func(*args, **kwargs)
65 return new_func
66
~/Research/Computing/miniconda3/envs/sci/lib/python3.6/site-packages/ESMF/api/grid.py in __init__(self, max_index, num_peri_dims, periodic_dim, pole_dim, coord_sys, coord_typekind, staggerloc, filename, filetype, reg_decomp, decompflag, is_sphere, add_corner_stagger, add_user_area, add_mask, varname, coord_names, tilesize, regDecompPTile, name)
451 # Add coordinates if a staggerloc is specified
452 if staggerloc is not None:
--> 453 self.add_coords(staggerloc=staggerloc, from_file=from_file)
454
455 # Add items if they are specified, this is done after the
~/Research/Computing/miniconda3/envs/sci/lib/python3.6/site-packages/ESMF/api/grid.py in add_coords(self, staggerloc, coord_dim, from_file)
797
798 # and now for Python
--> 799 self._allocate_coords_(stagger, from_file=from_file)
800
801 # set the staggerlocs to be done
~/Research/Computing/miniconda3/envs/sci/lib/python3.6/site-packages/ESMF/api/grid.py in _allocate_coords_(self, stagger, localde, from_file)
1022 if (self.ndims == self.rank) or (self.ndims == 0):
1023 for xyz in range(self.rank):
-> 1024 self._link_coord_buffer_(xyz, stagger, localde)
1025 # and this way if we have 1d coordinates
1026 elif self.ndims < self.rank:
~/Research/Computing/miniconda3/envs/sci/lib/python3.6/site-packages/ESMF/api/grid.py in _link_coord_buffer_(self, coord_dim, stagger, localde)
1072 lb, ub = ESMP_GridGetCoordBounds(self, staggerloc=stagger, localde=localde)
1073
-> 1074 gridCoordP = ndarray_from_esmf(data, self.type, ub-lb)
1075
1076 # alias the coordinates to a grid property
~/Research/Computing/miniconda3/envs/sci/lib/python3.6/site-packages/ESMF/util/esmpyarray.py in ndarray_from_esmf(data, dtype, shape)
37
38 esmfarray = np.ndarray(tuple(shape[:]), constants._ESMF2PythonType[dtype],
---> 39 buffer, order="F")
40
41 return esmfarray
TypeError: buffer is too small for requested array