HXL-style tag version: https://github.com/fireindark707/Python-Schema-Matching/tree/hxl_tag
HDXSM Datasets: https://drive.google.com/file/d/1wPXFS1ZLi6Xf7_aRZU1Jw_-dJrggs1Ox/view?usp=sharing
A python tool using XGboost and sentence-transformers to perform schema matching task on tables. Support multi-language column names and instances matching and can be used without column names. Both csv and json file type are supported.
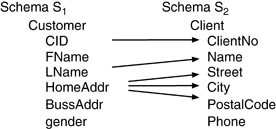
Schema matching is the problem of finding potential associations between elements (most often attributes or relations) of two schemas. source
- numpy==1.19.5
- pandas==1.1.5
- nltk==3.6.5
- python-dateutil==2.8.2
- sentence-transformers==2.1.0
- xgboost==1.5.2
- strsimpy==0.2.1
pip install schema-matching
from schema_matching import schema_matching
df_pred,df_pred_labels,predicted_pairs = schema_matching("Test Data/QA/Table1.json","Test Data/QA/Table2.json")
print(df_pred)
print(df_pred_labels)
for pair_tuple in predicted_pairs:
print(pair_tuple)
- df_pred: Predict value matrix, pd.DataFrame.
- df_pred_labels: Predict label matrix, pd.DataFrame.
- predicted_pairs: Predict label == 1 column pairs, in tuple format.
- table1_pth: Path to your first csv, json or jsonl file.
- table2_pth: Path to your second csv, json or jsonl file.
- threshold: Threshold, you can use this parameter to specify threshold value, suggest 0.9 for easy matching(column name very similar). Default value is calculated from training data, which is around 0.15-0.2. This value is used for difficult matching(column name masked or very different).
- strategy: Strategy, there are three options: "one-to-one", "one-to-many" and "many-to-many". "one-to-one" means that one column can only be matched to one column. "one-to-many" means that columns in Table1 can only be matched to one column in Table2. "many-to-many" means that there is no restrictions. Default is "many-to-many".
- model_pth: Path to trained model folder, which must contain at least one pair of ".model" file and ".threshold" file. You don't need to specify this parameter.
See Data format in Training Data and Test Data folders. You need to put mapping.txt, Table1.csv and Table2.csv in new folders under Training Data. For Test Data, mapping.txt is not needed.
python relation_features.py
python train.py
Example:
python cal_column_similarity.py -p Test\ Data/self -m /model/2022-04-12-12-06-32 -s one-to-one
python cal_column_similarity.py -p Test\ Data/authors -m /model/2022-04-12-12-06-32-11 -t 0.9
Parameters:
- -p: Path to test data folder, must contain "Table1.csv" and "Table2.csv" or "Table1.json" and "Table2.json".
- -m: Path to trained model folder, which must contain at least one pair of ".model" file and ".threshold" file.
- -t: Threshold, you can use this parameter to specify threshold value, suggest 0.9 for easy matching(column name very similar). Default value is calculated from training data, which is around 0.15-0.2. This value is used for difficult matching(column name masked or very different).
- -s: Strategy, there are three options: "one-to-one", "one-to-many" and "many-to-many". "one-to-one" means that one column can only be matched to one column. "one-to-many" means that columns in Table1 can only be matched to one column in Table2. "many-to-many" means that there is no restrictions. Default is "many-to-many".
Output:
- similarity_matrix_label.csv: Labels(0,1) for each column pairs.
- similarity_matrix_value.csv: Average of raw values computed by all the xgboost models.
Features: "is_url","is_numeric","is_date","is_string","numeric:mean", "numeric:min", "numeric:max", "numeric:variance","numeric:cv", "numeric:unique/len(data_list)", "length:mean", "length:min", "length:max", "length:variance","length:cv", "length:unique/len(data_list)", "whitespace_ratios:mean","punctuation_ratios:mean","special_character_ratios:mean","numeric_ratios:mean", "whitespace_ratios:cv","punctuation_ratios:cv","special_character_ratios:cv","numeric_ratios:cv", "colname:bleu_score", "colname:edit_distance","colname:lcs","colname:tsm_cosine", "colname:one_in_one", "instance_similarity:cosine"
- tsm_cosine: Cosine similarity of column names computed by sentence-transformers using "paraphrase-multilingual-mpnet-base-v2". Support multi-language column names matching.
- instance_similarity:cosine: Select 20 instances each string column and compute its mean embedding using sentence-transformers. Cosine similarity is computed by each pairs.
- Average Precision: 0.755
- Average Recall: 0.829
- Average F1: 0.766
Average Confusion Matrix:
| Negative(Truth) | Positive(Truth) | |
|---|---|---|
| Negative(pred) | 0.94343111 | 0.05656889 |
| Positive(pred) | 0.17135417 | 0.82864583 |
Data: https://github.com/fireindark707/Schema_Matching_XGboost/tree/main/Test%20Data/self
| title | text | summary | keywords | url | country | language | domain | name | timestamp | |
|---|---|---|---|---|---|---|---|---|---|---|
| col1 | 1(FN) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| col2 | 0 | 1(TP) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| col3 | 0 | 0 | 1(TP) | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| words | 0 | 0 | 0 | 1(TP) | 0 | 0 | 0 | 0 | 0 | 0 |
| link | 0 | 0 | 0 | 0 | 1(TP) | 0 | 0 | 0 | 0 | 0 |
| col6 | 0 | 0 | 0 | 0 | 0 | 1(TP) | 0 | 0 | 0 | 0 |
| lang | 0 | 0 | 0 | 0 | 0 | 0 | 1(TP) | 0 | 0 | 0 |
| col8 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1(TP) | 0 | 0 |
| website | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0(FN) | 0 |
| col10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1(TP) |
F1 score: 0.889
@software{fireinfark707_Schema_Matching_by_2022,
author = {fireinfark707},
license = {MIT},
month = {4},
title = {{Schema Matching by XGboost}},
url = {https://github.com/fireindark707/Schema_Matching_XGboost},
year = {2022}
}