The repository for all the datasets and projects related with data responsibly.
dataresponsibly / datasynthesizer Goto Github PK
View Code? Open in Web Editor NEWLicense: MIT License
License: MIT License
Your paper, DataSynthesizer: Privacy-Preserving Synthetic Datasets, says in section 3.1.3:
When invoked in independent attribute mode, DataDescriber performs frequency-based estimation of the unconditioned probability distributions of the attributes. The distribution is captured by bar charts for categorical attributes, and by histograms for numerical attributes. Precision of the histograms can be refined by a parameter named histogram size, which represents the number of bins and is set to 20 by default.
After checking the code for DataDescriber, I confirmed that this statement is also true for running in correlated attribute mode. Where does the figure 20 come from? Was this an arbitrary choice, or was there some motivation for this specific value?
Might I suggest that you use a variation of the Freedman–Diaconis rule instead? Or Scott's, or Sturges'? (More info here.)I believe this will lead to significant improvements in performance.
To demonstrate this, here's plots of two attributes from my dataset: age of person, and date of measured age (expressed as an integer count of days since 1900-01-01), with histogram_size = 20:
The synthesised data doesn't look so good.
However, if I use a variation of the Freedman-Diaconis rule to choose the number of bins (8), this is what I get:
Which looks much better!
Code for calculating number of bins for a 1D histogram can be got from SciPy
numpy.histogram
Hi,
Thank you so much for this! It's been a life saver. I got your model to run on one of my datasets, but I ran into a problem with higher degrees. With k = 2 and k = 3 models on my dataset, the code ran without bugs at several epsilons up to 2.5, but with k = 4 and higher, for all epsilons, this runs:
================ Constructing Bayesian Network (BN) ================
Adding ROOT accrued_holidays
Adding attribute org
Adding attribute office
Adding attribute start_date
Adding attribute bonus
Adding attribute birth_date
Adding attribute salary
Adding attribute title
Adding attribute gender
========================== BN constructed ==========================
But then the cell just freezes there until keyError (6,5,0,0) occurs
Hi, firstly, I haven't said it so far, but thanks for creating and maintaining DataSynthesizer! It's a useful tool.
DataDescriber creates a value missing_rate in the attribute descriptions. I was wondering what your thoughts are on making use of these values in DataGenerator along with the distribution bins which are already used.
My use case is pretty simple, I want to create a synthesised data set for non-production use which is as representative of the original data set as possible. Two extremes of the problem I'm having:
In some instances where it's more important for me, I have addressed this in pre and post-processing steps myself, but as DataDescriber collects this metric, I was wondering if it would be reasonable to implement this in DataSynthesizer itself, perhaps as an option passed to the relevant generator methods.
Cheers!
First spotted this error with my own data, but it is easy to replicate this behaviour with DataSynthesize Usage (correlated attribute mode).ipynb and the default adult_ssn data.
The Notebook states:
# A parameter in differential privacy.
# It roughtly means that removing one tuple will change the probability of any output by at most exp(epsilon).
# Set epsilon=0 to turn off differential privacy.
But setting epsilon = 0, instead of 0.1 (the default) makes DataDescriber crash:
ssn
age
education
marital-status
relationship
sex
income
---------------------------------------------------------------------------
ZeroDivisionError Traceback (most recent call last)
<ipython-input-18-62c183a34a81> in <module>()
2 describer.describe_dataset_in_correlated_attribute_mode(input_data, epsilon=epsilon, k=degree_of_bayesian_network,
3 attribute_to_is_categorical=categorical_attributes,
----> 4 attribute_to_is_candidate_key=candidate_keys)
5 describer.save_dataset_description_to_file(description_file)
~\OneDrive - EPAM\Documents\TR-data-masking\DataSynthesizer\DataDescriber.py in describe_dataset_in_correlated_attribute_mode(self, dataset_file, k, epsilon, attribute_to_datatype, attribute_to_is_categorical, attribute_to_is_candidate_key, seed)
121 self.describe_dataset_in_independent_attribute_mode(dataset_file, epsilon, attribute_to_datatype,
122 attribute_to_is_categorical, attribute_to_is_candidate_key,
--> 123 seed)
124 self.encoded_dataset = self.encode_dataset_into_binning_indices()
125 if self.encoded_dataset.shape[1] < 2:
~\OneDrive - EPAM\Documents\TR-data-masking\DataSynthesizer\DataDescriber.py in describe_dataset_in_independent_attribute_mode(self, dataset_file, epsilon, attribute_to_datatype, attribute_to_is_categorical, attribute_to_is_candidate_key, seed)
87 self.convert_input_dataset_into_a_dict_of_columns()
88 self.infer_domains()
---> 89 self.inject_laplace_noise_into_distribution_per_attribute(epsilon)
90 # record attribute information in json format
91 self.dataset_description['attribute_description'] = {}
~\OneDrive - EPAM\Documents\TR-data-masking\DataSynthesizer\DataDescriber.py in inject_laplace_noise_into_distribution_per_attribute(self, epsilon)
246 for column in self.input_dataset_as_column_dict.values():
247 assert isinstance(column, AbstractAttribute)
--> 248 column.inject_laplace_noise(epsilon, num_attributes_in_BN)
249
250 def encode_dataset_into_binning_indices(self):
~\OneDrive - EPAM\Documents\TR-data-masking\DataSynthesizer\datatypes\AbstractAttribute.py in inject_laplace_noise(self, epsilon, num_valid_attributes)
52
53 def inject_laplace_noise(self, epsilon=0.1, num_valid_attributes=10):
---> 54 noisy_scale = num_valid_attributes / (epsilon * self.data.size)
55 laplace_noises = np.random.laplace(0, scale=noisy_scale, size=len(self.distribution_probabilities))
56 noisy_distribution = np.asarray(self.distribution_probabilities) + laplace_noises
ZeroDivisionError: division by zero
because of line 54.
Setting epsilon to zero doesn't turn off differential privacy; I'm guessing there should have been a check for the value of zero before executing any function involved differential privacy.
I've followed the installation instructions and am attempting to run "DataSynthesizer Usage (correlated attribute mode).ipynb" I get the following error when running Step 4:
---------------------------------------------------------------------------
TypeError Traceback (most recent call last)
<ipython-input-10-4621f1d6556b> in <module>()
1 generator = DataGenerator()
----> 2 generator.generate_dataset_in_correlated_attribute_mode(num_tuples_to_generate, description_file)
3 generator.save_synthetic_data(synthetic_data)
~/DataSynthesizer/DataSynthesizer/DataGenerator.py in generate_dataset_in_correlated_attribute_mode(self, n, description_file, seed)
68
69 if attr in self.encoded_dataset:
---> 70 self.synthetic_dataset[attr] = column.sample_values_from_binning_indices(self.encoded_dataset[attr])
71 elif attr in candidate_keys:
72 self.synthetic_dataset[attr] = column.generate_values_as_candidate_key(n)
~/DataSynthesizer/DataSynthesizer/datatypes/IntegerAttribute.py in sample_values_from_binning_indices(self, binning_indices)
18
19 def sample_values_from_binning_indices(self, binning_indices):
---> 20 column = super().sample_values_from_binning_indices(binning_indices)
21 column[~column.isnull()] = column[~column.isnull()].astype(int)
22 return column
~/DataSynthesizer/DataSynthesizer/datatypes/AbstractAttribute.py in sample_values_from_binning_indices(self, binning_indices)
96 def sample_values_from_binning_indices(self, binning_indices):
97 """ Convert binning indices into values in domain. Used by both independent and correlated attribute mode. """
---> 98 return binning_indices.apply(lambda x: self.uniform_sampling_within_a_bin(x))
99
100 def uniform_sampling_within_a_bin(self, binning_index):
~/anaconda3/lib/python3.6/site-packages/pandas/core/series.py in apply(self, func, convert_dtype, args, **kwds)
3190 else:
3191 values = self.astype(object).values
-> 3192 mapped = lib.map_infer(values, f, convert=convert_dtype)
3193
3194 if len(mapped) and isinstance(mapped[0], Series):
pandas/_libs/src/inference.pyx in pandas._libs.lib.map_infer()
~/DataSynthesizer/DataSynthesizer/datatypes/AbstractAttribute.py in <lambda>(x)
96 def sample_values_from_binning_indices(self, binning_indices):
97 """ Convert binning indices into values in domain. Used by both independent and correlated attribute mode. """
---> 98 return binning_indices.apply(lambda x: self.uniform_sampling_within_a_bin(x))
99
100 def uniform_sampling_within_a_bin(self, binning_index):
~/DataSynthesizer/DataSynthesizer/datatypes/AbstractAttribute.py in uniform_sampling_within_a_bin(self, binning_index)
106 bins = self.distribution_bins.copy()
107 bins.append(2 * bins[-1] - bins[-2])
--> 108 return uniform(bins[binning_index], bins[binning_index + 1])
TypeError: list indices must be integers or slices, not float
The other notebooks work just fine, so it seems my environment is setup correctly, but there is a bug with the correlated attribute mode.
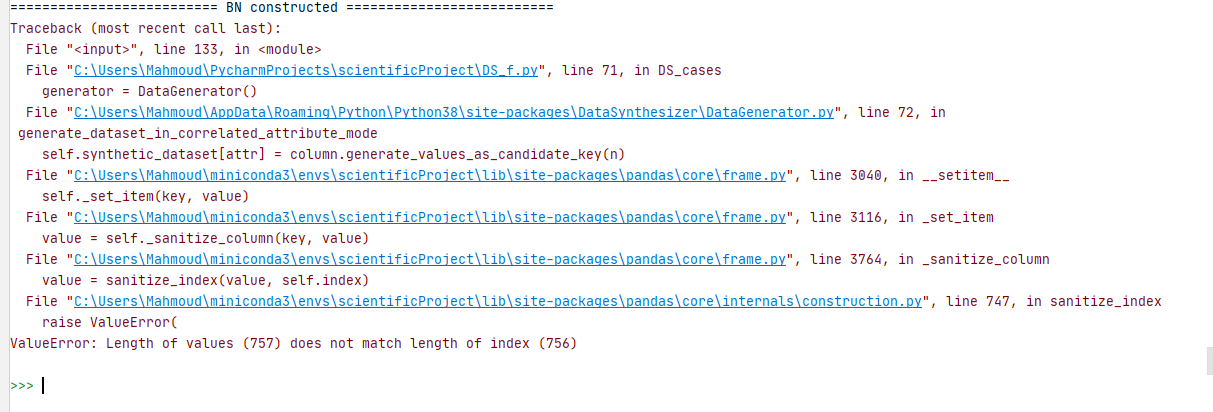
I'm trying to use the Data generator in correlated attribute mode.I tried with many datasets and everything works fine. However, for some datasets, I'm getting the following error when I run the DataGenerator:
ValueError: Length of values (757) does not match length of index (756)
Note that the DataDescriber works fine without raising an error. However, for the datasets that I weren't able to generate syntehtic data, I checked their description file, and in all of them, the number of attributes in the Bayesian network is less than the number of attributes in the whole datasets.
I have a CSV with ~20 columns, 3 of which are unique identifiers. DataSynthesizer seems to be tripping up on these 3 columns with the error below. What's the expected behaviour when trying to include UUIDs (or similar) in the synthesise? The field is not labelled as categorical and is of datatype String.
describer = DataDescriber()
generator = DataGenerator()
describer.describe_dataset_in_random_mode(
dataset_file='input.csv',
attribute_to_datatype=attribute_to_datatype,
attribute_to_is_categorical=attribute_is_categorical,
)
describer.save_dataset_description_to_file('description.csv'),
)
generator.generate_dataset_in_random_mode(
n,
'output.csv',
)Traceback (most recent call last):
File "synthesise/synthesise.py", line 106, in <module>
main()
File "synthesise/synthesise.py", line 86, in main
generator.generate_dataset_in_correlated_attribute_mode(
File "/Users/raids/.pyenv/versions/data-synthesizer/lib/python3.8/site-packages/DataSynthesizer/DataGenerator.py", line 72, in generate_dataset_in_correlated_attribute_mode
self.synthetic_dataset[attr] = column.generate_values_as_candidate_key(n)
File "/Users/raids/.pyenv/versions/data-synthesizer/lib/python3.8/site-packages/DataSynthesizer/datatypes/StringAttribute.py", line 52, in generate_values_as_candidate_key
length = np.random.randint(self.min, self.max)
File "mtrand.pyx", line 745, in numpy.random.mtrand.RandomState.randint
File "_bounded_integers.pyx", line 1254, in numpy.random._bounded_integers._rand_int64
ValueError: low >= highLet me know if you need further info or want me to try anything out.
Thanks
Describing a dataset in independent attribute mode can fail during infer_distribution() for String attributes if a subset of the values could be inferred as numerical. sort_index() is called on a pd.Series which results in the following TypeError:
Traceback (most recent call last):
File "main.py", line 76, in <module>
args.func(args)
File "main.py", line 40, in synthesise
d = synthesise(mode=mode, sample=sample)
File "/Users/raids/synth/synthesise/synthesiser.py", line 104, in synthesise
self.describer.describe_dataset_in_independent_attribute_mode(
File "/Users/raids/.pyenv/versions/data-synthesizer/lib/python3.8/site-packages/DataSynthesizer/DataDescriber.py", line 123, in describe_dataset_in_independent_attribute_mode
column.infer_distribution()
File "/Users/raids/.pyenv/versions/data-synthesizer/lib/python3.8/site-packages/DataSynthesizer/datatypes/StringAttribute.py", line 49, in infer_distribution
distribution.sort_index(inplace=True)
File "/Users/raids/.pyenv/versions/data-synthesizer/lib/python3.8/site-packages/pandas/core/series.py", line 3156, in sort_index
indexer = nargsort(
File "/Users/raids/.pyenv/versions/data-synthesizer/lib/python3.8/site-packages/pandas/core/sorting.py", line 274, in nargsort
indexer = non_nan_idx[non_nans.argsort(kind=kind)]
TypeError: '<' not supported between instances of 'str' and 'float'I ran into this with a specific string attribute and patched infer_distribution() with a single line; see line with comment below:
def infer_distribution(self):
if self.is_categorical:
distribution = self.data_dropna.value_counts()
for value in set(self.distribution_bins) - set(distribution.index):
distribution[value] = 0
distribution.index = distribution.index.map(str) # patch to fix index type
distribution.sort_index(inplace=True)
self.distribution_probabilities = utils.normalize_given_distribution(distribution)
self.distribution_bins = np.array(distribution.index)
else:
distribution = np.histogram(self.data_dropna_len, bins=self.histogram_size)
self.distribution_bins = distribution[1][:-1]
self.distribution_probabilities = utils.normalize_given_distribution(distribution[0])Happy to fork and raise a PR for this change and the other attributes if you think it's an appropriate fix, not sure if the other data types would require anything like this, though, nor non-categorical attributes (when it falls into the else above.
Let me know what you think and if you need anything from my side.
Cheers
When I run describe_dataset_in_correlated_attribute_mod, the first value of the first DataFrame column of string objects is being read as the month. For a given record, the attributes are
I believe the sensitivity of the conditional distributions is wrong and as a result you're injecting too little noise to the conditional distributions in the correlated attribute mode with enabled epsilon.
In Algorithm 1 in PrivBayes, the stated sensitivity is 2 / n_tuples because the Laplace noise is added to the joint probability (as the probability is already materialised). However, you apply the Laplace noise to the counts (as opposed to a probability) which have sensitivity of 2 (and not 2 / n_tuples). Only later you normalise the counts to materialise the conditional probability. As a result, you're injecting too little noise to the conditional probabilities.
Code excerpts from DataSynthesizer/lib/PrivBayes.py:
# Applying Laplace to the counts:
noise_para = laplace_noise_parameter(k, num_attributes, num_tuples, epsilon)
# with noise_para = 2 * (num_attributes - k) / (num_tuples * epsilon)
laplace_noises = np.random.laplace(0, scale=noise_para, size=stats.index.size)
stats['count'] += laplace_noises
# Normalising counts to a probability distribution:
dist = normalize_given_distribution(stats_sub['count']).tolist()
In order to inject the correct amount of noise 2 * (num_attributes - k) / (num_tuples * epsilon) should be changed to 2 * (num_attributes - k) / epsilon.
PS: thanks to @sofiane87 and @bristena-op for the helpful discussions and clarifications.
Apologies if I'm just being dense though is there documentation on the different modes enabled by DataSynthesizer? Aka random, independent attribute, correlated. Guessing has to do with assumptions with the data generating process for the dataset in question though would love to dive deeper.
PS love the goals with this tool!
Greetings!
I'm trying to understand your paper and implementation. I've noticed that the more you increase epsilon, the less noise will be generated. In order to understand if that is the expected behavior, I looked into your paper and PrivBayes paper (and, also, a Java implementation) and everyone seems to say that the scale of the noise is given by:
4 * (n_cols - k) / (n_rows * epsilon)
But the definition of differential privacy implies that if epsilon gets closer to 0, there won't be any difference for the query output between the original and synthetic datasets. Am I getting something wrong?
Thanks in advance!
I am running DataSynthesizer on a toy data set with 8 columns and ~10,000 rows. Only 2 of the columns are correlated so I expect to see this correlation in the synthetic data set when running in correlated mode.
I noticed that every time I run (with all parameters constant), the Bayesian netwok that is generated is different - I think this is a result of the greedy algorithm converging to a different solution. The two correlated columns are connected with a parent/child relationship only sometimes. When they are not connected the correlation in the synthetic data is close to zero as expected. I tried to change k from 2 to 4 but the same thing happens and the runtime increases a lot.
As a result, I also cannot compare how DataSynthesiser performs (in terms of correlations) when varying one of the parameter (e.g. epsilon) because when the right relationships are not included the results are completely different.
Is there a way to force the greedy algorithm to connect two of the columns with a parent/child relationship? Or to pass a pre-defined graph, allowing the user to define all relationships? In that case the only task for the algorithm would be the inference of the probability values.
Also, why is the greedy algorithm unable to detect the relationship between the two variables, given that it is the only pair that is correlated in the entire dataset?
After cd into webUI/, installing all the dependencies locally and running django manage.py runserver, an error occurs!
Here's the error stack trace:
Unhandled exception in thread started by <function check_errors.<locals>.wrapper at 0x10df840d0>
Traceback (most recent call last):
File "/Users/me/projects/workforce-data-initiative/DataSynthesizer/webUI/venv/lib/python3.6/site-packages/django/utils/autoreload.py", line 227, in wrapper
fn(*args, **kwargs)
File "/Users/me/projects/workforce-data-initiative/DataSynthesizer/webUI/venv/lib/python3.6/site-packages/django/core/management/commands/runserver.py", line 117, in inner_run
autoreload.raise_last_exception()
File "/Users/me/projects/workforce-data-initiative/DataSynthesizer/webUI/venv/lib/python3.6/site-packages/django/utils/autoreload.py", line 250, in raise_last_exception
six.reraise(*_exception)
File "/Users/me/projects/workforce-data-initiative/DataSynthesizer/webUI/venv/lib/python3.6/site-packages/django/utils/six.py", line 685, in reraise
raise value.with_traceback(tb)
File "/Users/me/projects/workforce-data-initiative/DataSynthesizer/webUI/venv/lib/python3.6/site-packages/django/utils/autoreload.py", line 227, in wrapper
fn(*args, **kwargs)
File "/Users/me/projects/workforce-data-initiative/DataSynthesizer/webUI/venv/lib/python3.6/site-packages/django/__init__.py", line 27, in setup
apps.populate(settings.INSTALLED_APPS)
File "/Users/me/projects/workforce-data-initiative/DataSynthesizer/webUI/venv/lib/python3.6/site-packages/django/apps/registry.py", line 85, in populate
app_config = AppConfig.create(entry)
File "/Users/me/projects/workforce-data-initiative/DataSynthesizer/webUI/venv/lib/python3.6/site-packages/django/apps/config.py", line 120, in create
mod = import_module(mod_path)
File "/Users/me/projects/workforce-data-initiative/DataSynthesizer/webUI/venv/lib/python3.6/importlib/__init__.py", line 126, in import_module
return _bootstrap._gcd_import(name[level:], package, level)
File "<frozen importlib._bootstrap>", line 994, in _gcd_import
File "<frozen importlib._bootstrap>", line 971, in _find_and_load
File "<frozen importlib._bootstrap>", line 941, in _find_and_load_unlocked
File "<frozen importlib._bootstrap>", line 219, in _call_with_frames_removed
File "<frozen importlib._bootstrap>", line 994, in _gcd_import
File "<frozen importlib._bootstrap>", line 971, in _find_and_load
File "<frozen importlib._bootstrap>", line 953, in _find_and_load_unlocked
ModuleNotFoundError: No module named 'dataflow'
Hello,
I am trying to generate synthetic data where one column comprises timestamp.
I am using random mode. Everything seems fine. The program throws no errors. But the timestamps are generated in float.
INPUT:
timestamp
--
17-10-1999
13-12-1999
22-03-2000
23-03-2000
25-03-2000
05-05-2000
15-05-2000
23-05-2000
OUTPUT:
timestamp
--
84.53577
53.62745
68.02702
60.91776
9.8478
9.202759
5.596583
8.653249
In Python 3.11, describe_dataset_in_correlated_attribute_mode raises ValueError. And in Python 3.10, the same code with the same versions of dependencies works correctly.
At the same time, describe_dataset_in_independent_attribute_mode and describe_dataset_in_random_mode work correctly in Python 3.11.
Pandas version is 1.5.3, and not the latest 2.0.3, as describe_dataset_in_correlated_attribute_mode additionally doesn't work with Pandas 2.0.3 (I will write a separate issue on that later).
from DataSynthesizer.DataDescriber import DataDescriber
describer = DataDescriber()
describer.describe_dataset_in_correlated_attribute_mode(dataset_file=input_data, k=2, epsilon=0)
describer.save_dataset_description_to_file(description_file)When the code is ran, following happens:
Traceback:
ValueError Traceback (most recent call last)
Cell In[22], line 8
6 describer = DataDescriber()
7 #TODO k parameter
----> 8 describer.describe_dataset_in_correlated_attribute_mode(dataset_file=input_data,
9 k=2,
10 epsilon=0)
11 #seed=random_state,
12 #attribute_to_is_categorical=categorical_attributes)
13 describer.save_dataset_description_to_file(description_file)
File ~\.virtualenvs\DataSynthesizerTest311\Lib\site-packages\DataSynthesizer\DataDescriber.py:177, in DataDescriber.describe_dataset_in_correlated_attribute_mode(self, dataset_file, k, epsilon, attribute_to_datatype, attribute_to_is_categorical, attribute_to_is_candidate_key, categorical_attribute_domain_file, numerical_attribute_ranges, seed)
174 if self.df_encoded.shape[1] < 2:
175 raise Exception("Correlated Attribute Mode requires at least 2 attributes(i.e., columns) in dataset.")
--> 177 self.bayesian_network = greedy_bayes(self.df_encoded, k, epsilon / 2, seed=seed)
178 self.data_description['bayesian_network'] = self.bayesian_network
179 self.data_description['conditional_probabilities'] = construct_noisy_conditional_distributions(
180 self.bayesian_network, self.df_encoded, epsilon / 2)
File ~\.virtualenvs\DataSynthesizerTest311\Lib\site-packages\DataSynthesizer\lib\PrivBayes.py:145, in greedy_bayes(dataset, k, epsilon, seed)
142 attr_to_is_binary = {attr: dataset[attr].unique().size <= 2 for attr in dataset}
144 print('================ Constructing Bayesian Network (BN) ================')
--> 145 root_attribute = random.choice(dataset.columns)
146 V = [root_attribute]
147 rest_attributes = list(dataset.columns)
File C:\Python311\Lib\random.py:369, in Random.choice(self, seq)
367 def choice(self, seq):
368 """Choose a random element from a non-empty sequence."""
--> 369 if not seq:
370 raise IndexError('Cannot choose from an empty sequence')
371 return seq[self._randbelow(len(seq))]
File ~\.virtualenvs\DataSynthesizerTest311\Lib\site-packages\pandas\core\indexes\base.py:3188, in Index.__nonzero__(self)
3186 @final
3187 def __nonzero__(self) -> NoReturn:
-> 3188 raise ValueError(
3189 f"The truth value of a {type(self).__name__} is ambiguous. "
3190 "Use a.empty, a.bool(), a.item(), a.any() or a.all()."
3191 )
ValueError: The truth value of a Index is ambiguous. Use a.empty, a.bool(), a.item(), a.any() or a.all().Hello,
I'm trying to synthesizer datasets with high number of attributes (above 50). However, the data synthesis process is taking too long (multiple hours).
Is there a way we can speed up the process? Can we run it on the GPU?
Paste the command(s) you ran and the output.
If there was a crash, please include the traceback here.
Thanks for the good work!!
Am looking at the bayesian creation code and have questions regarding line 153-155 and line 111in the PrivBayes.py code:
num_parents = min(len(V), k)
tasks = [(child, V, num_parents, split, dataset) for child, split in
product(rest_attributes, range(len(V) - num_parents + 1))]
What is the rationale behind generating a list of combinations with different split points for each attribute in the rest_attributes list?
It seems like the worker function code can account for all the combinations of attribute and parents pairs in line 111 , just by looking at the entire V for each attribute, instead of iterating all possible V[split:] for each attribute.
Hi,
In a float or int field, it appears that the pandas lib treats them as string fields rather than flat with null value.
Is there anyway to force float, either in the UI or in the pandas read method?
While trying to convert a string data from the input to a DateTime DataSynthesizer format, something is going wrong:
Traceback (most recent call last):
File "/Users/stijngeuens/Google Drive/Digipolis/syntheticCRS-P/generator.py", line 32, in
attribute_to_is_categorical=attribute_is_categorical)
File "/Users/stijngeuens/Google Drive/Digipolis/syntheticCRS-P/DataSynthesizer/DataDescriber.py", line 172, in describe_dataset_in_correlated_attribute_mode
seed)
File "/Users/stijngeuens/Google Drive/Digipolis/syntheticCRS-P/DataSynthesizer/DataDescriber.py", line 120, in describe_dataset_in_independent_attribute_mode
seed=seed)
File "/Users/stijngeuens/Google Drive/Digipolis/syntheticCRS-P/DataSynthesizer/DataDescriber.py", line 98, in describe_dataset_in_random_mode
column.infer_domain()
File "/Users/stijngeuens/Google Drive/Digipolis/syntheticCRS-P/DataSynthesizer/datatypes/DateTimeAttribute.py", line 44, in infer_domain
self.min = float(self.data_dropna.min())
ValueError: could not convert string to float: '1904-02-22'
I have data that contains a dozen or so columns that contain categorical variables with many distinct values. When I try to run in correlated attribute mode, line 162 in PrivBayes.py results in MemoryError: I run out of RAM. Here's the line:
full_space = pd.DataFrame(columns=attributes, data=list(product(*stats.index.levels)))
Without knowing much about the workings of the code, it looks like it's taking cartesian product over all the columns in my data so that DataDescriber can learn the joint distributions of subsets of (categorical) data selected by the Bayesian network finding routine, some of which have thousands of distinct values, and consequently the line above is trying to generate massive tables that are maxing-out RAM. That's just a guess.
Other than running in independent attribute mode, do you have any recommendations for how to proceed when data contains many categorical variables with many distinct values? It's often not possible to merge values by binning them into a smaller number of distinct values. For example, my PlaceOfBirth column contains (city, country) combinations that result in a huge number of values, and it's neither meaningful to separate the city and country information, nor bin the data, and I'd like to retain the relationships to other columns in the data, such as Passport or Citizenship, because they are meaningful. This is just an example of a general problem that I've encountered. One might often encounter data like this.
Is there a technical workaround, or is there some best practice that you can recommend for dealing with this situation --- how can I use your software optimally? Anything documented?
Hello,
I am using DataSynthesizer to generate synthetic data for research purposes. I've been using this package for moths and it works perfectly with small datasets. However, when I use a bigger dataset, especially higher number of columns, time problem rises. A single dataset(with 71236 instances and 52) took more than 18 hours to be synthesized on a 64 core machine(degree_of_bayesian_network =0 in this case) .
I also tried to decrease the degree_of_bayesian_network , by assigning it to 2 instead of the default 0. Although the quality of the synthesized data decreases, Time decreases , but it's still taking too long.
What do you suggest to do? Is there a better way you recommend to approach bigger datasets?
Paste the command(s) you ran and the output.
If there was a crash, please include the traceback here.
I use Google Colab.
My input dataset has a column, which contains 2 distinct DateTime values: "2020-09-14", "2020-09-16".
describe_dataset_in_correlated_attribute_mode works correctly.
But when I run generate_dataset_in_correlated_attribute_mode, following error appears (I also included full traceback):
ValueError: invalid literal for int() with base 10: '2020-09-14'
I did some investigation, and the relevant part of description file looks like this (I also included full description file):
"dateTime": {
"name": "dateTime",
"data_type": "DateTime",
"is_categorical": true,
"is_candidate_key": false,
"min": 1600041600.0,
"max": 1600214400.0,
"missing_rate": 0.0,
"distribution_bins": [
"2020-09-14",
"2020-09-16"
],
"distribution_probabilities": [
0.6587202007528231,
0.341279
]
},
For some reason, "min" and "max" are timestamps, and "distribution_bins" are strings. It's suspicious.
It's the only place in the description flie where the datetime literals "2020-09-14" and "2020-09-16" appear.
ValueError Traceback (most recent call last)
in ()
1 generator = DataGenerator()
----> 2 generator.generate_dataset_in_correlated_attribute_mode(num_tuples_to_generate, description_file)
3 generator.save_synthetic_data(synthetic_data)
9 frames
/usr/local/lib/python3.7/dist-packages/DataSynthesizer/DataGenerator.py in generate_dataset_in_correlated_attribute_mode(self, n, description_file, seed)
68
69 if attr in self.encoded_dataset:
---> 70 self.synthetic_dataset[attr] = column.sample_values_from_binning_indices(self.encoded_dataset[attr])
71 elif attr in candidate_keys:
72 self.synthetic_dataset[attr] = column.generate_values_as_candidate_key(n)
/usr/local/lib/python3.7/dist-packages/DataSynthesizer/datatypes/DateTimeAttribute.py in sample_values_from_binning_indices(self, binning_indices)
83 def sample_values_from_binning_indices(self, binning_indices):
84 column = super().sample_values_from_binning_indices(binning_indices)
---> 85 column[~column.isnull()] = column[~column.isnull()].astype(int)
86 return column
/usr/local/lib/python3.7/dist-packages/pandas/core/generic.py in astype(self, dtype, copy, errors)
5813 else:
5814 # else, only a single dtype is given
-> 5815 new_data = self._mgr.astype(dtype=dtype, copy=copy, errors=errors)
5816 return self._constructor(new_data).finalize(self, method="astype")
5817
/usr/local/lib/python3.7/dist-packages/pandas/core/internals/managers.py in astype(self, dtype, copy, errors)
416
417 def astype(self: T, dtype, copy: bool = False, errors: str = "raise") -> T:
--> 418 return self.apply("astype", dtype=dtype, copy=copy, errors=errors)
419
420 def convert(
/usr/local/lib/python3.7/dist-packages/pandas/core/internals/managers.py in apply(self, f, align_keys, ignore_failures, **kwargs)
325 applied = b.apply(f, **kwargs)
326 else:
--> 327 applied = getattr(b, f)(**kwargs)
328 except (TypeError, NotImplementedError):
329 if not ignore_failures:
/usr/local/lib/python3.7/dist-packages/pandas/core/internals/blocks.py in astype(self, dtype, copy, errors)
589 values = self.values
590
--> 591 new_values = astype_array_safe(values, dtype, copy=copy, errors=errors)
592
593 new_values = maybe_coerce_values(new_values)
/usr/local/lib/python3.7/dist-packages/pandas/core/dtypes/cast.py in astype_array_safe(values, dtype, copy, errors)
1307
1308 try:
-> 1309 new_values = astype_array(values, dtype, copy=copy)
1310 except (ValueError, TypeError):
1311 # e.g. astype_nansafe can fail on object-dtype of strings
/usr/local/lib/python3.7/dist-packages/pandas/core/dtypes/cast.py in astype_array(values, dtype, copy)
1255
1256 else:
-> 1257 values = astype_nansafe(values, dtype, copy=copy)
1258
1259 # in pandas we don't store numpy str dtypes, so convert to object
/usr/local/lib/python3.7/dist-packages/pandas/core/dtypes/cast.py in astype_nansafe(arr, dtype, copy, skipna)
1172 # work around NumPy brokenness, #1987
1173 if np.issubdtype(dtype.type, np.integer):
-> 1174 return lib.astype_intsafe(arr, dtype)
1175
1176 # if we have a datetime/timedelta array of objects
/usr/local/lib/python3.7/dist-packages/pandas/_libs/lib.pyx in pandas._libs.lib.astype_intsafe()
ValueError: invalid literal for int() with base 10: '2020-09-14'
The code which I ran:
generator = DataGenerator()
generator.generate_dataset_in_correlated_attribute_mode(num_tuples_to_generate, description_file)
generator.save_synthetic_data(synthetic_data)
Description file:
fns_fiscal_generated_description.zip
The function get_noisy_distribution_of_attributes only gets a partial distribution. This bug was introduced in commit 1abe702. Here is the relevant code as it appears in master (currently commit be8b65a):
full_space = None
for item in grouper_it(products, 1000000):
if full_space is None:
full_space = DataFrame(columns=attributes, data=list(item))
else:
data_frame_append = DataFrame(columns=attributes, data=list(item))
full_space.append(data_frame_append)In particular, full_space.append does not modify full_space; instead, it returns a new object. (This seems to be true for all versions of pandas.) As a result, full_space does not store all of the intended rows but, rather, only at most the first 1000000.
DataDescriber does not handle well bool dtypes in the source dataset. When the CSV file has columns with only TRUE and FALSE as values, pandas reads such columns as bool dtype (not object) and, when inferring types, the code ends up in checking them as dates and fails.
The source dataset is the telco-customer-churn dataset from Kaggle, after being imported in Google BigQuery and exported back to CSV, generating those TRUE and FALSE values instead of Yes and No. Below is my code:
from DataSynthesizer.DataDescriber import DataDescriber
from DataSynthesizer.DataGenerator import DataGenerator
from DataSynthesizer.ModelInspector import ModelInspector
from DataSynthesizer.lib.utils import read_json_file, display_bayesian_network
import pandas as pd
# input dataset
input_data = "./out/from_bq.csv" # this CSV file has columns with TRUE and FALSE value which get read by pandas as bool dtype
mode = 'correlated_attribute_mode'
# location of two output files
description_file = f'./out/{mode}/description.json'
synthetic_data = f'./out/{mode}/synthetic_data.csv'
# An attribute is categorical if its domain size is less than this threshold.
threshold_value = 20
# list of dicsrete columns and primary key
categorical_columns = ["gender", "Partner", "Dependents", "PhoneService", "MultipleLines", "InternetService", "OnlineSecurity", "OnlineBackup", "DeviceProtection", "TechSupport", "StreamingTV", "StreamingMovies", "Contract", "PaperlessBilling", "PaymentMethod", "TotalCharges", "Churn"]
primary_key_column = "customerID"
# specify categorical attributes
categorical_attributes = {}
for column in categorical_columns:
categorical_attributes[column] = True
# specify which attributes are candidate keys of input dataset.
candidate_keys = {primary_key_column: True}
# A parameter in Differential Privacy. It roughly means that removing a row in the input dataset will not
# change the probability of getting the same output more than a multiplicative difference of exp(epsilon).
# Increase epsilon value to reduce the injected noises. Set epsilon=0 to turn off differential privacy.
epsilon = 0
# The maximum number of parents in Bayesian network, i.e., the maximum number of incoming edges.
degree_of_bayesian_network = 2
# Number of tuples generated in synthetic dataset.
num_tuples_to_generate = 1000
# build the Bayesian Network
describer = DataDescriber(category_threshold=threshold_value)
describer.describe_dataset_in_correlated_attribute_mode(dataset_file="./out/from_bq.csv",
epsilon=epsilon,
k=degree_of_bayesian_network,
attribute_to_is_categorical=categorical_attributes,
attribute_to_is_candidate_key=candidate_keys)
# save the output
describer.save_dataset_description_to_file(description_file)
Here is the output:
---------------------------------------------------------------------------
AttributeError Traceback (most recent call last)
/tmp/ipykernel_4364/1321006366.py in <module>
46 k=degree_of_bayesian_network,
47 attribute_to_is_categorical=categorical_attributes,
---> 48 attribute_to_is_candidate_key=candidate_keys)
49
50 # save the output
/opt/conda/lib/python3.7/site-packages/DataSynthesizer/DataDescriber.py in describe_dataset_in_correlated_attribute_mode(self, dataset_file, k, epsilon, attribute_to_datatype, attribute_to_is_categorical, attribute_to_is_candidate_key, categorical_attribute_domain_file, numerical_attribute_ranges, seed)
170 categorical_attribute_domain_file,
171 numerical_attribute_ranges,
--> 172 seed)
173 self.df_encoded = self.encode_dataset_into_binning_indices()
174 if self.df_encoded.shape[1] < 2:
/opt/conda/lib/python3.7/site-packages/DataSynthesizer/DataDescriber.py in describe_dataset_in_independent_attribute_mode(self, dataset_file, epsilon, attribute_to_datatype, attribute_to_is_categorical, attribute_to_is_candidate_key, categorical_attribute_domain_file, numerical_attribute_ranges, seed)
118 categorical_attribute_domain_file,
119 numerical_attribute_ranges,
--> 120 seed=seed)
121
122 for column in self.attr_to_column.values():
/opt/conda/lib/python3.7/site-packages/DataSynthesizer/DataDescriber.py in describe_dataset_in_random_mode(self, dataset_file, attribute_to_datatype, attribute_to_is_categorical, attribute_to_is_candidate_key, categorical_attribute_domain_file, numerical_attribute_ranges, seed)
85 self.attr_to_is_candidate_key = attribute_to_is_candidate_key
86 self.read_dataset_from_csv(dataset_file)
---> 87 self.infer_attribute_data_types()
88 self.analyze_dataset_meta()
89 self.represent_input_dataset_by_columns()
/opt/conda/lib/python3.7/site-packages/DataSynthesizer/DataDescriber.py in infer_attribute_data_types(self)
213 # Sample 20 values to test its data_type.
214 samples = column_dropna.sample(20, replace=True)
--> 215 if all(samples.map(is_datetime)):
216 self.attr_to_datatype[attr] = DataType.DATETIME
217 else:
/opt/conda/lib/python3.7/site-packages/pandas/core/series.py in map(self, arg, na_action)
3980 dtype: object
3981 """
-> 3982 new_values = super()._map_values(arg, na_action=na_action)
3983 return self._constructor(new_values, index=self.index).__finalize__(
3984 self, method="map"
/opt/conda/lib/python3.7/site-packages/pandas/core/base.py in _map_values(self, mapper, na_action)
1158
1159 # mapper is a function
-> 1160 new_values = map_f(values, mapper)
1161
1162 return new_values
pandas/_libs/lib.pyx in pandas._libs.lib.map_infer()
/opt/conda/lib/python3.7/site-packages/DataSynthesizer/datatypes/DateTimeAttribute.py in is_datetime(value)
19 'dec', 'december'}
20
---> 21 value_lower = value.lower()
22 if (value_lower in weekdays) or (value_lower in months):
23 return False
AttributeError: 'bool' object has no attribute 'lower'
Im trying to install DataSynthesizer on Anaconda for using on jupiter notebook
When i pip install DataSynthesizer i get this error:
ERROR: Could not find a version that satisfies the requirement DataSynthesizer (from versions: none)
ERROR: No matching distribution found for DataSynthesizer
I also try with conda install DataSynthesizer and get the next message
PackagesNotFoundError: The following packages are not available from current channels:
- datasynthesizer
I'm trying out your data synthesizer on correlated_attribute_mode using your example on python notebook.
I've generated data using epsilon values 0.0001 and 1 to compare how different the generated data is but they are exactly the same. Is this normal?
When I try to upload dataset to the Django app I get the following error:
Currently this repo is without any explicit license terms. Under what terms is the software made available?
Should DataSynthesizer in either independent_attribute_mode or correlated_attribute_mode support floating point numbers?
If so, it seems to have a problem - if not, guess I missed that in the documentation!
I have a small sample dataset - attached in csv format (but ending in .txt due to github upload requirements) that causes an error when it gets to the column with float values.
sample_data_no_dates.txt
Here's the error I receive when I use the attached csv as the input_data:
---------------------------------------------------------------------------
ValueError Traceback (most recent call last)
<ipython-input-12-4621f1d6556b> in <module>()
1 generator = DataGenerator()
----> 2 generator.generate_dataset_in_correlated_attribute_mode(num_tuples_to_generate, description_file)
3 generator.save_synthetic_data(synthetic_data)
~/DataSynthesizer/DataSynthesizer/DataGenerator.py in generate_dataset_in_correlated_attribute_mode(self, n, description_file, seed)
74 print('candidate keys:', column.generate_values_as_candidate_key(n))
75 print('self.synthetic_dataset:', self.synthetic_dataset)
---> 76 self.synthetic_dataset[attr] = column.generate_values_as_candidate_key(n)
77 else:
78 # for attributes not in BN or candidate keys, use independent attribute mode.
~/anaconda3/lib/python3.6/site-packages/pandas/core/frame.py in __setitem__(self, key, value)
3114 else:
3115 # set column
-> 3116 self._set_item(key, value)
3117
3118 def _setitem_slice(self, key, value):
~/anaconda3/lib/python3.6/site-packages/pandas/core/frame.py in _set_item(self, key, value)
3189
3190 self._ensure_valid_index(value)
-> 3191 value = self._sanitize_column(key, value)
3192 NDFrame._set_item(self, key, value)
3193
~/anaconda3/lib/python3.6/site-packages/pandas/core/frame.py in _sanitize_column(self, key, value, broadcast)
3386
3387 # turn me into an ndarray
-> 3388 value = _sanitize_index(value, self.index, copy=False)
3389 if not isinstance(value, (np.ndarray, Index)):
3390 if isinstance(value, list) and len(value) > 0:
~/anaconda3/lib/python3.6/site-packages/pandas/core/series.py in _sanitize_index(data, index, copy)
3996
3997 if len(data) != len(index):
-> 3998 raise ValueError('Length of values does not match length of ' 'index')
3999
4000 if isinstance(data, ABCIndexClass) and not copy:
ValueError: Length of values does not match length of index
When I use the DataDescriber I get a file not found error. I would like to save the dataset description in a .json file that does not exist yet. I would like DataSynthesizer to create this file for me. Isn't there an option to add a small function that checks the filepath and then creates the empty file if the specified path/file is non-existent? Similar to these two lines of code on this post from StackOverflow. Then I do not have to create empty files before running the programming.
I suggest adding these two lines in this function:
DataSynthesizer/DataSynthesizer/DataDescriber.py
Lines 301 to 303 in 0bae2c1
Happy to hear your thoughts on this, thanks!
Hi,
I have a really small clinical dataset with 176 rows and 80 columns. Some of the variables are considered to be categorical and other continuous; all the NaNs have been replaced with -1 in integer/float and "" in string.
I filled the configuration dictionaries attr_to_datatype, categorical_attributes and candidate_keys with the variables in my dataset.
I'm trying to reproduce the example notebook in correlated attribute mode (I know for sure there are some correlations in my dataset) without touching anything else except for the settings mentioned above.
It seems like nothing is working properly because none of the data generated follows the real distribution of the original dataset but it looks like they're following some sort of uniform distribution. Here you can see some examples:
I can't find any solution to my problem and I've been trying every kind of settings combination but none seems to work correctly.
Maybe I'm missing something or there might be some problem in the generation process.
What do you suggest to do ? Thank you.
When i run the webui i get an error.
This is what i did:
PYTHONPATH=../DataSynthesizer python manage.py runserver
Watching for file changes with StatReloader
Performing system checks...
System check identified no issues (0 silenced).
You have 17 unapplied migration(s). Your project may not work properly until you apply the migrations for app(s): admin, auth, contenttypes, sessions.
Run 'python manage.py migrate' to apply them.
Then i go to http://127.0.0.1:8000/ and go to tools->Data Synthesizer tool
and i get a Gateway 502 error'?
I figured trying again annd running python manage.py migrate would fix the issue but instead i get ModuleNotFoundError: No module named 'DataGenerator'...
I followed the installation instructions as described in readme.
In DateTimeAttribute.py, line 65:
timestamps = self.data_dropna.map(lambda x: parse(x).timestamp())
timestamp() results in a crash for dates earlier than 1970:
Traceback (most recent call last):
File "C:\Users\ANDREW~1\AppData\Local\Temp\RtmpuMDjSt\chunk-code-2b143b894699.txt", line 131, in <module>
describer.describe_dataset_in_correlated_attribute_mode(input_data, epsilon = epsilon, k = degree_of_bayesian_network, attribute_to_is_categorical = categorical_attributes, attribute_to_is_candidate_key = candidate_keys)
File ".\DataSynthesizer\DataDescriber.py", line 123, in describe_dataset_in_correlated_attribute_mode
seed)
File ".\DataSynthesizer\DataDescriber.py", line 88, in describe_dataset_in_independent_attribute_mode
self.infer_domains()
File ".\DataSynthesizer\DataDescriber.py", line 242, in infer_domains
column.infer_domain(self.input_dataset[column.name])
File ".\DataSynthesizer\datatypes\DateTimeAttribute.py", line 56, in infer_domain
timestamps = self.data_dropna.map(lambda x: parse(x).timestamp())
File "C:\Users\Andrew_Lowe\AppData\Local\Continuum\Anaconda3\lib\site-packages\pandas\core\series.py", line 2354, in map
new_values = map_f(values, arg)
File "pandas/_libs/src/inference.pyx", line 1521, in pandas._libs.lib.map_infer
File ".\DataSynthesizer\datatypes\DateTimeAttribute.py", line 56, in <lambda>
timestamps = self.data_dropna.map(lambda x: parse(x).timestamp())
OSError: [Errno 22] Invalid argument
This is apparently a known Python bug: see this
Stack Overflow post.
If the timestamp is out of the range of values supported by the platform C
localtime()orgmtime()functions,datetime.fromtimestamp()may raise an exception like you're seeing. On Windows platform, this range can sometimes be restricted to years in 1970 through 2038. I have never seen this problem on a Linux system.
The same problem seems to occur with timestamp(); I tried this from a Python command prompt:
>>> from dateutil.parser import parse
>>> parse('19/04/1979').timestamp()
293320800.0
>>> parse('19/04/1969').timestamp()
Traceback (most recent call last):
File "<stdin>", line 1, in <module>
OSError: [Errno 22] Invalid argument
If you're not seeing this behaviour, the SO post hints that Windows systems are affected, but not Linux.
Is there way to replace the translation from dates to timestamps, and vice versa, with code that works for dates earlier than 1970-01-01?
Hey, we are thinking of using your system in our project. It seems like only categorical attributes are calculated by the privbayes algorithm. The numeric values such as integer/float does not go through privbayes algorithm.
Say I have a dataset with 4 variables(2 nonsensible, 2 sensible), of which I only want to synthsize the last two(sensible variables). By What I understand from DataDescriber, the only way to not synthesize a variable is to set it as a candidate_keys which will simply ennumerate each row of the data.
If I treat the first two variables(nonsensible variables) just like the others, I can run into the problem, that the attribute-parent tuples from the BN are in a problematic order. The sensible variable could be a parent of the nonsensible variable. Of course I would like to have it the other way around, so that the first 2 nonsensible variables can provide information (being so to speak somehow treated as "predictors") for sampling the sensible variables.
Maybe I am missing some obvious way to do partial synthesis?
A declarative, efficient, and flexible JavaScript library for building user interfaces.
🖖 Vue.js is a progressive, incrementally-adoptable JavaScript framework for building UI on the web.
TypeScript is a superset of JavaScript that compiles to clean JavaScript output.
An Open Source Machine Learning Framework for Everyone
The Web framework for perfectionists with deadlines.
A PHP framework for web artisans
Bring data to life with SVG, Canvas and HTML. 📊📈🎉
JavaScript (JS) is a lightweight interpreted programming language with first-class functions.
Some thing interesting about web. New door for the world.
A server is a program made to process requests and deliver data to clients.
Machine learning is a way of modeling and interpreting data that allows a piece of software to respond intelligently.
Some thing interesting about visualization, use data art
Some thing interesting about game, make everyone happy.
We are working to build community through open source technology. NB: members must have two-factor auth.
Open source projects and samples from Microsoft.
Google ❤️ Open Source for everyone.
Alibaba Open Source for everyone
Data-Driven Documents codes.
China tencent open source team.