Get metadata, fulltexts or fulltext URLs of papers matching a search query using any of the following APIs:
- EuropePMC
- IEEE
- ArXiv
- Crossref (metadata, no fulltext)
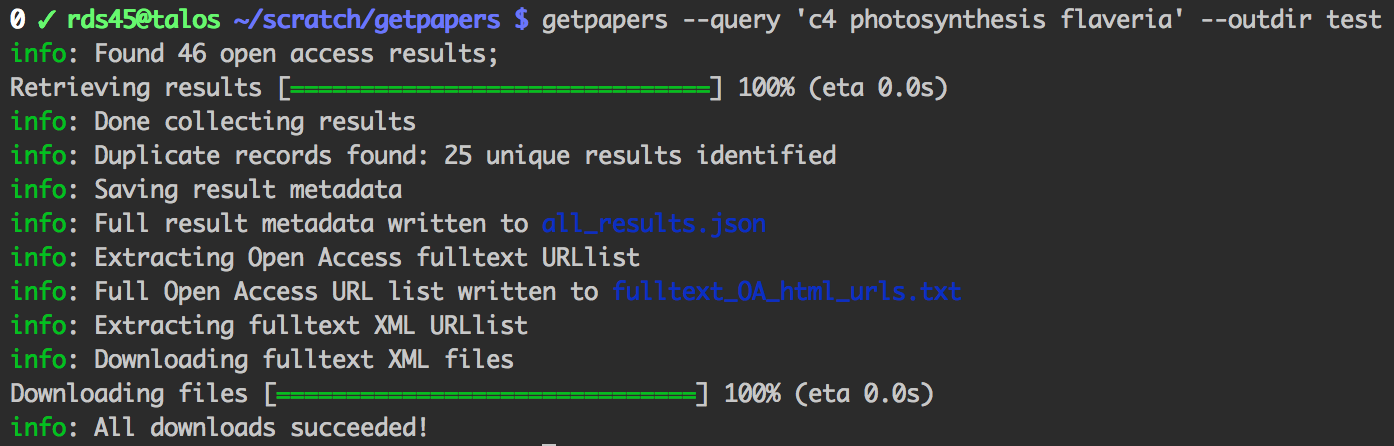
getpapers can fetch article metadata, fulltexts (PDF or XML), and supplementary materials. It's designed for use in content mining, but you may find it useful for quickly acquiring large numbers of papers for reading, or for bibliometrics.
Please follow these cross-platform instructions
$ npm install --global getpapersUse getpapers --help to see the command-line help:
-h, --help output usage information
-V, --version output the version number
-q, --query <query> search query (required)
-o, --outdir <path> output directory (required - will be created if not found)
--api <name> API to search [eupmc, crossref, ieee, arxiv] (default: eupmc)
-x, --xml download fulltext XMLs if available
-p, --pdf download fulltext PDFs if available
-s, --supp download supplementary files if available
-t, --minedterms download text-mined terms if available
-l, --loglevel <level> amount of information to log (silent, verbose, info*, data, warn, error, or debug)
-a, --all search all papers, not just open access
-n, --noexecute report how many results match the query, but don't actually download anything
-f, --logfile <filename> save log to specified file in output directory as well as printing to terminal
-k, --limit <int> limit the number of hits and downloads
--filter <filter object> filter by key value pair, passed straight to the crossref api only
-r, --restart restart file downloads after failure
By default, getpapers uses the EuropePMC API.
Each API has its own query format. Usage guides are provided on our wiki:
- EuropePMC query format
- IEEE query format
- ArXiv query format
- Crossref API [Crossref docs]
Copyright (c) 2014 Shuttleworth Foundation Licensed under the MIT license
- The remote site may timeout or hang (we have found that if EPMC gets a query with no results it will timeout).
- Be careful not to download the whole site. use the
-koption to limit downloads (this should be a default).