Companion Packages:
Table of Contents
Documentation + Code Examples
Expand to view content
Automated Machine Learning - In my view, AutoML should consist of functions to help make professional model development and operationalization more efficient. The functions in this package are there to help no matter which part of the ML lifecycle you are working on. The functions in this package have been tested across a variety of industries and have consistently outperformed competing methods.
Supervised Learning - Currently, I'm utilizing CatBoost, LightGBM, XGBoost, and H2O for all of the automated Machine Learning related functions. GPU's can be utilized with CatBoost, LightGBM, and XGBoost, while those and the H2O models can all utilize 100% of CPU. Multi-armed bandit grid tuning is available for CatBoost, LightGBM, and XGBoost models, which utilize the concept of randomized probability matching, which is detailed in the R pacakge "bandit". My choice of included ML algorithms in the package is based on previous success when compared against other algorithms on real world use cases, the additional utilities these packages offer aside from accurate predictions, their ability to work on big data, and the fact that they're available in both R and Python which makes managing multiple languages a little more seamless in a professional setting.
Documentation - Each exported function in the package has a help file and can be viewed in your RStudio session, e.g.
?Rodeo::ModelDataPrep. Many of them come with examples coded up in the help files (at the bottom) that you can run to get a feel for how to set the parameters. There's also a listing of exported functions by category with code examples at the bottom of this readme. You can also jump into the R folder here to dig into the source code.
Overall process: Typically, I go to the warehouse to get all of my base features and then I run through all the relevant feature engineering functions in this package. Personally, I set up templates for features engineering, model training optimization, and model scoring (including feature engineering for scoring). I collect all relevant metdata in a list that is shared across templates and as a result, I never have to touch the model scoring template, which makes operationalize and maintenace a breeze. I can simply list out the columns of interest, which feature engineering functions I want to utilize, and then I simply kick off some command line scripts and everything else is automatically managed.
The Description File is designed to require only the minimum number of packages to install AutoQuant. However, in order to utilize most of the functions in the package, you'll have to install additional libraries. I set it up this way on purpose. You don't need to install every single possible dependency if you are only interested in using a few of the functions. For example, if you only want to use CatBoost then install the catboost package and forget about the h2o, xgboost, and lightgbm packages. This is one of the primary benefits of not hosting an R package on cran, as they require dependencies to be part of the Imports section on the Description File, which subsequently requires users to have all dependencies installed in order to install the package.
The minimal set of packages that need to be installed are below. The full list can be found by expanding the section (Expand to view content).
- bit64
- data.table
- doParallel
- foreach
- lubridate
- timeDate
# Core pacakges
if(!("data.table" %in% rownames(installed.packages()))) install.packages("data.table"); print("data.table")
if(!("collapse" %in% rownames(installed.packages()))) install.packages("collapse"); print("collapse")
if(!("bit64" %in% rownames(installed.packages()))) install.packages("bit64"); print("bit64")
if(!("devtools" %in% rownames(installed.packages()))) install.packages("devtools"); print("devtools")
if(!("doParallel" %in% rownames(installed.packages()))) install.packages("doParallel"); print("doParallel")
if(!("foreach" %in% rownames(installed.packages()))) install.packages("foreach"); print("foreach")
if(!("lubridate" %in% rownames(installed.packages()))) install.packages("lubridate"); print("lubridate")
if(!("timeDate" %in% rownames(installed.packages()))) install.packages("timeDate"); print("timeDate")
# AutoQuant
devtools::install_github('AdrianAntico/AutoQuant', upgrade = FALSE, dependencies = FALSE, force = TRUE)Additional Packages to Install
XGBoost and LightGBM can be used with GPU. However, their installation is much more involved than CatBoost, which comes with GPU capabilities simply by installing their package. The installation instructions for them below is for the CPU version only. Refer to each's home page for instructions for installing for GPU.
# Install Dependencies----
if(!("devtools" %in% rownames(installed.packages()))) install.packages("devtools"); print("devtools")
# Core pacakges
if(!("data.table" %in% rownames(installed.packages()))) install.packages("data.table"); print("data.table")
if(!("collapse" %in% rownames(installed.packages()))) install.packages("collapse"); print("collapse")
if(!("bit64" %in% rownames(installed.packages()))) install.packages("bit64"); print("bit64")
if(!("devtools" %in% rownames(installed.packages()))) install.packages("devtools"); print("devtools")
if(!("doParallel" %in% rownames(installed.packages()))) install.packages("doParallel"); print("doParallel")
if(!("foreach" %in% rownames(installed.packages()))) install.packages("foreach"); print("foreach")
if(!("lubridate" %in% rownames(installed.packages()))) install.packages("lubridate"); print("lubridate")
if(!("timeDate" %in% rownames(installed.packages()))) install.packages("timeDate"); print("timeDate")
# Additional dependencies for specific use cases
if(!("combinat" %in% rownames(installed.packages()))) install.packages("combinat"); print("combinat")
if(!("DBI" %in% rownames(installed.packages()))) install.packages("DBI"); print("DBI")
if(!("e1071" %in% rownames(installed.packages()))) install.packages("e1071"); print("e1071")
if(!("fBasics" %in% rownames(installed.packages()))) install.packages("fBasics"); print("fBasics")
if(!("forecast" %in% rownames(installed.packages()))) install.packages("forecast"); print("forecast")
if(!("fpp" %in% rownames(installed.packages()))) install.packages("fpp"); print("fpp")
if(!("ggplot2" %in% rownames(installed.packages()))) install.packages("ggplot2"); print("ggplot2")
if(!("gridExtra" %in% rownames(installed.packages()))) install.packages("gridExtra"); print("gridExtra")
if(!("itertools" %in% rownames(installed.packages()))) install.packages("itertools"); print("itertools")
if(!("MLmetrics" %in% rownames(installed.packages()))) install.packages("MLmetrics"); print("MLmetrics")
if(!("nortest" %in% rownames(installed.packages()))) install.packages("nortest"); print("nortest")
if(!("pROC" %in% rownames(installed.packages()))) install.packages("pROC"); print("pROC")
if(!("RColorBrewer" %in% rownames(installed.packages()))) install.packages("RColorBrewer"); print("RColorBrewer")
if(!("recommenderlab" %in% rownames(installed.packages()))) install.packages("recommenderlab"); print("recommenderlab")
if(!("RPostgres" %in% rownames(installed.packages()))) install.packages("RPostgres"); print("RPostgres")
if(!("Rfast" %in% rownames(installed.packages()))) install.packages("Rfast"); print("Rfast")
if(!("scatterplot3d" %in% rownames(installed.packages()))) install.packages("scatterplot3d"); print("scatterplot3d")
if(!("stringr" %in% rownames(installed.packages()))) install.packages("stringr"); print("stringr")
if(!("tsoutliers" %in% rownames(installed.packages()))) install.packages("tsoutliers"); print("tsoutliers")
if(!("xgboost" %in% rownames(installed.packages()))) install.packages("xgboost"); print("xgboost")
if(!("lightgbm" %in% rownames(installed.packages()))) install.packages("lightgbm"); print("lightgbm")
if(!("regmedint" %in% rownames(installed.packages()))) install.packages("regmedint"); print("regmedint")
for(pkg in c("RCurl","jsonlite")) if (! (pkg %in% rownames(installed.packages()))) { install.packages(pkg) }
install.packages("h2o", type = "source", repos = (c("http://h2o-release.s3.amazonaws.com/h2o/latest_stable_R")))
devtools::install_github('catboost/catboost', subdir = 'catboost/R-package')
# Dependencies for ML Reports
if(!("reactable" %in% rownames(installed.packages()))) install.packages("reactable"); print("reactable")
devtools::install_github('AdrianAntico/prettydoc', upgrade = FALSE, dependencies = FALSE, force = TRUE)
# And lastly, AutoQuant
devtools::install_github('AdrianAntico/AutoQuant', upgrade = FALSE, dependencies = FALSE, force = TRUE)The most common issue some users are having when trying to install AutoQuant is the installation of the catboost package dependency. Since catboost is not on CRAN it can only be installed through GitHub. To install catboost without error (and consequently install AutoQuant without error), try running this line of code first, then restart your R session, then re-run the 2-step installation process above. (Reference):
If you're still having trouble submit an issue and I'll work with you to get it installed.
# Method for on premise servers
options(devtools.install.args = c("--no-multiarch", "--no-test-load"))
install.packages("https://github.com/catboost/catboost/releases/download/<version>/catboost-R-Windows-<version>.tgz", repos = NULL, type = "source", INSTALL_opts = c("--no-multiarch", "--no-test-load"))
# Method for azure machine learning Designer pipelines
## catboost
install.packages("https://github.com/catboost/catboost/releases/download/<version>/catboost-R-Windows-<version>.tgz", repos = NULL, type = "source", INSTALL_opts = c("--no-multiarch", "--no-test-load"))
## AutoQuant
install.packages("https://github.com/AdrianAntico/AutoQuant/archive/refs/tags/<version>.tar.gz", repos = NULL, type = "source", INSTALL_opts = c("--no-multiarch", "--no-test-load"))Expand to view content
click to expand
Regression Description
The Auto_Regression() models handle a multitude of tasks. In order:
- Convert your data to data.table format for faster processing
- Transform your target variable using the best normalization method based on the
AutoTransformationCreate()function - Create train, validation, and test data, utilizing the
AutoDataPartition()function, if you didn't supply those directly to the function - Consoldate columns that are used for modeling and what metadata you want returned in your test data with predictions
- Dichotomize categorical variables (for
AutoXGBoostRegression()) and save the factor levels for scoring in a way that guarentees consistency across training, validation, and test data sets, utilizing theDummifyDT()function - Save the final modeling column names for reference
- Handles the data conversion to the appropriate modeling type, such as CatBoost, H2O, and XGBoost
- Multi-armed bandit hyperparameter tuning using randomized probability matching, if you choose to grid tune
- Loop through the grid-tuning process, building N models
- Collect the evaluation metrics for each grid tune run
- Identify the best model of the set of models built in the grid tuning search
- Save the hyperparameters from the winning grid tuned model
- Build the final model based on the best model from the grid tuning model search (I remove each model after evaluation metrics are generated in the grid tune to avoid memory overflow)
- Back-transform your predictions based on the best transformation used earlier in the process
- Collect evaluation metrics based on performance on test data (based on back-transformed data)
- Store the final predictions with the associated test data and other columns you want included in that set
- Save your transformation metadata for recreating them in a scoring process
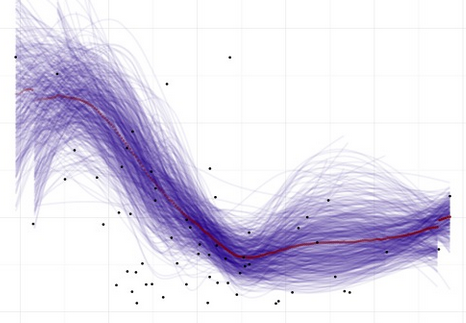
- Build out and save an Evaluation Calibration Line Plot and Evaluation Calibration Box-Plot, using the
EvalPlot()function - Generate and save Variable Importance
- Generate and save Partital Dependence Calibration Line Plots and Partital Dependence Calibration Box-Plots, using the
ParDepPlots()function - Return all the objects generated in a named list for immediate use and evaluation
CatBoost Example
# Create some dummy correlated data
data <- AutoQuant::FakeDataGenerator(
Correlation = 0.85,
N = 10000,
ID = 2,
ZIP = 0,
AddDate = FALSE,
Classification = FALSE,
MultiClass = FALSE)
# Run function
TestModel <- AutoQuant::AutoCatBoostRegression(
# GPU or CPU and the number of available GPUs
TrainOnFull = FALSE,
task_type = 'GPU',
NumGPUs = 1,
DebugMode = FALSE,
# Metadata args
OutputSelection = c('Importances', 'EvalPlots', 'EvalMetrics', 'Score_TrainData'),
ModelID = 'Test_Model_1',
model_path = normalizePath('./'),
metadata_path = normalizePath('./'),
SaveModelObjects = FALSE,
SaveInfoToPDF = FALSE,
ReturnModelObjects = TRUE,
# Data args
data = data,
ValidationData = NULL,
TestData = NULL,
TargetColumnName = 'Adrian',
FeatureColNames = names(data)[!names(data) %in%
c('IDcol_1', 'IDcol_2','Adrian')],
PrimaryDateColumn = NULL,
WeightsColumnName = NULL,
IDcols = c('IDcol_1','IDcol_2'),
TransformNumericColumns = 'Adrian',
Methods = c('BoxCox', 'Asinh', 'Asin', 'Log',
'LogPlus1', 'Sqrt', 'Logit'),
# Model evaluation
eval_metric = 'RMSE',
eval_metric_value = 1.5,
loss_function = 'RMSE',
loss_function_value = 1.5,
MetricPeriods = 10L,
NumOfParDepPlots = ncol(data)-1L-2L,
# Grid tuning args
PassInGrid = NULL,
GridTune = FALSE,
MaxModelsInGrid = 30L,
MaxRunsWithoutNewWinner = 20L,
MaxRunMinutes = 60*60,
BaselineComparison = 'default',
# ML args
langevin = FALSE,
diffusion_temperature = 10000,
Trees = 1000,
Depth = 9,
L2_Leaf_Reg = NULL,
RandomStrength = 1,
BorderCount = 128,
LearningRate = NULL,
RSM = 1,
BootStrapType = NULL,
GrowPolicy = 'SymmetricTree',
model_size_reg = 0.5,
feature_border_type = 'GreedyLogSum',
sampling_unit = 'Object',
subsample = NULL,
score_function = 'Cosine',
min_data_in_leaf = 1)XGBoost Example
# Create some dummy correlated data
data <- AutoQuant::FakeDataGenerator(
Correlation = 0.85,
N = 1000,
ID = 2,
ZIP = 0,
AddDate = FALSE,
Classification = FALSE,
MultiClass = FALSE)
# Run function
TestModel <- AutoQuant::AutoXGBoostRegression(
# GPU or CPU
TreeMethod = 'hist',
NThreads = parallel::detectCores(),
LossFunction = 'reg:squarederror',
# Metadata args
OutputSelection = c('Importances', 'EvalPlots', 'EvalMetrics', 'Score_TrainData'),
model_path = normalizePath("./"),
metadata_path = NULL,
ModelID = "Test_Model_1",
EncodingMethod = "binary",
ReturnFactorLevels = TRUE,
ReturnModelObjects = TRUE,
SaveModelObjects = FALSE,
SaveInfoToPDF = FALSE,
DebugMode = FALSE,
# Data args
data = data,
TrainOnFull = FALSE,
ValidationData = NULL,
TestData = NULL,
TargetColumnName = "Adrian",
FeatureColNames = names(data)[!names(data) %in% c('IDcol_1','IDcol_2','Adrian')],
PrimaryDateColumn = NULL,
WeightsColumnName = NULL,
IDcols = c('IDcol_1','IDcol_2'),
TransformNumericColumns = 'Adrian',
Methods = c('Asinh','Asin','Log','LogPlus1','Sqrt','Logit'),
# Model evaluation args
eval_metric = 'rmse',
NumOfParDepPlots = 3L,
# Grid tuning args
PassInGrid = NULL,
GridTune = FALSE,
grid_eval_metric = 'r2',
BaselineComparison = 'default',
MaxModelsInGrid = 10L,
MaxRunsWithoutNewWinner = 20L,
MaxRunMinutes = 24L*60L,
Verbose = 1L,
# ML args
Trees = 50L,
eta = 0.05,
max_depth = 4L,
min_child_weight = 1.0,
subsample = 0.55,
colsample_bytree = 0.55)LightGBM Example
# Create some dummy correlated data
data <- AutoQuant::FakeDataGenerator(
Correlation = 0.85,
N = 1000,
ID = 2,
ZIP = 0,
AddDate = FALSE,
Classification = FALSE,
MultiClass = FALSE)
# Run function
TestModel <- AutoQuant::AutoLightGBMRegression(
# Metadata args
OutputSelection = c('Importances','EvalPlots','EvalMetrics','Score_TrainData'),
model_path = normalizePath('./'),
metadata_path = NULL,
ModelID = 'Test_Model_1',
NumOfParDepPlots = 3L,
EncodingMethod = 'credibility',
ReturnFactorLevels = TRUE,
ReturnModelObjects = TRUE,
SaveModelObjects = FALSE,
SaveInfoToPDF = FALSE,
DebugMode = FALSE,
# Data args
data = data,
TrainOnFull = FALSE,
ValidationData = NULL,
TestData = NULL,
TargetColumnName = 'Adrian',
FeatureColNames = names(data)[!names(data) %in% c('IDcol_1', 'IDcol_2','Adrian')],
PrimaryDateColumn = NULL,
WeightsColumnName = NULL,
IDcols = c('IDcol_1','IDcol_2'),
TransformNumericColumns = NULL,
Methods = c('Asinh','Asin','Log','LogPlus1','Sqrt','Logit'),
# Grid parameters
GridTune = FALSE,
grid_eval_metric = 'r2',
BaselineComparison = 'default',
MaxModelsInGrid = 10L,
MaxRunsWithoutNewWinner = 20L,
MaxRunMinutes = 24L*60L,
PassInGrid = NULL,
# Core parameters
# https://lightgbm.readthedocs.io/en/latest/Parameters.html#core-parameters
input_model = NULL, # continue training a model that is stored to file
task = 'train',
device_type = 'CPU',
NThreads = parallel::detectCores() / 2,
objective = 'regression',
metric = 'rmse',
boosting = 'gbdt',
LinearTree = FALSE,
Trees = 50L,
eta = NULL,
num_leaves = 31,
deterministic = TRUE,
# Learning Parameters
# https://lightgbm.readthedocs.io/en/latest/Parameters.html#learning-control-parameters
force_col_wise = FALSE,
force_row_wise = FALSE,
max_depth = NULL,
min_data_in_leaf = 20,
min_sum_hessian_in_leaf = 0.001,
bagging_freq = 0,
bagging_fraction = 1.0,
feature_fraction = 1.0,
feature_fraction_bynode = 1.0,
extra_trees = FALSE,
early_stopping_round = 10,
first_metric_only = TRUE,
max_delta_step = 0.0,
lambda_l1 = 0.0,
lambda_l2 = 0.0,
linear_lambda = 0.0,
min_gain_to_split = 0,
drop_rate_dart = 0.10,
max_drop_dart = 50,
skip_drop_dart = 0.50,
uniform_drop_dart = FALSE,
top_rate_goss = FALSE,
other_rate_goss = FALSE,
monotone_constraints = NULL,
monotone_constraints_method = 'advanced',
monotone_penalty = 0.0,
forcedsplits_filename = NULL, # use for AutoStack option; .json file
refit_decay_rate = 0.90,
path_smooth = 0.0,
# IO Dataset Parameters
# https://lightgbm.readthedocs.io/en/latest/Parameters.html#io-parameters
max_bin = 255,
min_data_in_bin = 3,
data_random_seed = 1,
is_enable_sparse = TRUE,
enable_bundle = TRUE,
use_missing = TRUE,
zero_as_missing = FALSE,
two_round = FALSE,
# Convert Parameters
convert_model = NULL,
convert_model_language = 'cpp',
# Objective Parameters
# https://lightgbm.readthedocs.io/en/latest/Parameters.html#objective-parameters
boost_from_average = TRUE,
alpha = 0.90,
fair_c = 1.0,
poisson_max_delta_step = 0.70,
tweedie_variance_power = 1.5,
lambdarank_truncation_level = 30,
# Metric Parameters (metric is in Core)
# https://lightgbm.readthedocs.io/en/latest/Parameters.html#metric-parameters
is_provide_training_metric = TRUE,
eval_at = c(1,2,3,4,5),
# Network Parameters
# https://lightgbm.readthedocs.io/en/latest/Parameters.html#network-parameters
num_machines = 1,
# GPU Parameters
# https://lightgbm.readthedocs.io/en/latest/Parameters.html#gpu-parameters
gpu_platform_id = -1,
gpu_device_id = -1,
gpu_use_dp = TRUE,
num_gpu = 1)H2O-GBM Example
# Create some dummy correlated data
data <- AutoQuant::FakeDataGenerator(
Correlation = 0.85,
N = 1000,
ID = 2,
ZIP = 0,
AddDate = FALSE,
Classification = FALSE,
MultiClass = FALSE)
# Run function
TestModel <- AutoQuant::AutoH2oGBMRegression(
# Compute management
MaxMem = {gc();paste0(as.character(floor(as.numeric(system("awk '/MemFree/ {print $2}' /proc/meminfo", intern=TRUE)) / 1000000)),"G")},
NThreads = max(1, parallel::detectCores()-2),
H2OShutdown = TRUE,
H2OStartUp = TRUE,
IfSaveModel = "mojo",
# Model evaluation
NumOfParDepPlots = 3,
# Metadata arguments:
model_path = normalizePath("./"),
metadata_path = file.path(normalizePath("./")),
ModelID = "FirstModel",
ReturnModelObjects = TRUE,
SaveModelObjects = FALSE,
SaveInfoToPDF = FALSE,
# Data arguments
data = data,
TrainOnFull = FALSE,
ValidationData = NULL,
TestData = NULL,
TargetColumnName = 'Adrian',
FeatureColNames = names(data)[!names(data) %in% c('IDcol_1','IDcol_2','Adrian')],
WeightsColumn = NULL,
TransformNumericColumns = NULL,
Methods = c('Asinh','Asin','Log','LogPlus1','Sqrt','Logit'),
# ML grid tuning args
GridTune = FALSE,
GridStrategy = "Cartesian",
MaxRuntimeSecs = 60*60*24,
StoppingRounds = 10,
MaxModelsInGrid = 2,
# Model args
Trees = 50,
LearnRate = 0.10,
LearnRateAnnealing = 1,
eval_metric = "RMSE",
Alpha = NULL,
Distribution = "poisson",
MaxDepth = 20,
SampleRate = 0.632,
ColSampleRate = 1,
ColSampleRatePerTree = 1,
ColSampleRatePerTreeLevel = 1,
MinRows = 1,
NBins = 20,
NBinsCats = 1024,
NBinsTopLevel = 1024,
HistogramType = "AUTO",
CategoricalEncoding = "AUTO")H2O-DRF Example
# Create some dummy correlated data
data <- AutoQuant::FakeDataGenerator(
Correlation = 0.85,
N = 1000,
ID = 2,
ZIP = 0,
AddDate = FALSE,
Classification = FALSE,
MultiClass = FALSE)
# Run function
TestModel <- AutoQuant::AutoH2oDRFRegression(
# Compute management
MaxMem = {gc();paste0(as.character(floor(as.numeric(system("awk '/MemFree/ {print $2}' /proc/meminfo", intern=TRUE)) / 1000000)),"G")},
NThreads = max(1L, parallel::detectCores() - 2L),
H2OShutdown = TRUE,
H2OStartUp = TRUE,
IfSaveModel = "mojo",
# Model evaluation:
eval_metric = "RMSE",
NumOfParDepPlots = 3,
# Metadata arguments:
model_path = normalizePath("./"),
metadata_path = NULL,
ModelID = "FirstModel",
ReturnModelObjects = TRUE,
SaveModelObjects = FALSE,
SaveInfoToPDF = FALSE,
# Data Args
data = data,
TrainOnFull = FALSE,
ValidationData = NULL,
TestData = NULL,
TargetColumnName = "Adrian",
FeatureColNames = names(data)[!names(data) %in% c("IDcol_1", "IDcol_2","Adrian")],
WeightsColumn = NULL,
TransformNumericColumns = NULL,
Methods = c("Asinh", "Asin", "Log", "LogPlus1", "Sqrt", "Logit"),
# Grid Tuning Args
GridStrategy = "Cartesian",
GridTune = FALSE,
MaxModelsInGrid = 10,
MaxRuntimeSecs = 60*60*24,
StoppingRounds = 10,
# ML Args
Trees = 50,
MaxDepth = 20,
SampleRate = 0.632,
MTries = -1,
ColSampleRatePerTree = 1,
ColSampleRatePerTreeLevel = 1,
MinRows = 1,
NBins = 20,
NBinsCats = 1024,
NBinsTopLevel = 1024,
HistogramType = "AUTO",
CategoricalEncoding = "AUTO")H2O-GLM Example
# Create some dummy correlated data
data <- AutoQuant::FakeDataGenerator(
Correlation = 0.85,
N = 1000,
ID = 2,
ZIP = 0,
AddDate = FALSE,
Classification = FALSE,
MultiClass = FALSE)
# Run function
TestModel <- AutoQuant::AutoH2oGLMRegression(
# Compute management
MaxMem = {gc();paste0(as.character(floor(as.numeric(system("awk '/MemFree/ {print $2}' /proc/meminfo", intern=TRUE)) / 1000000)),"G")},
NThreads = max(1, parallel::detectCores()-2),
H2OShutdown = TRUE,
H2OStartUp = TRUE,
IfSaveModel = "mojo",
# Model evaluation:
eval_metric = "RMSE",
NumOfParDepPlots = 3,
# Metadata arguments:
model_path = NULL,
metadata_path = NULL,
ModelID = "FirstModel",
ReturnModelObjects = TRUE,
SaveModelObjects = FALSE,
SaveInfoToPDF = FALSE,
# Data arguments:
data = data,
TrainOnFull = FALSE,
ValidationData = NULL,
TestData = NULL,
TargetColumnName = "Adrian",
FeatureColNames = names(data)[!names(data) %in% c("IDcol_1", "IDcol_2","Adrian")],
RandomColNumbers = NULL,
InteractionColNumbers = NULL,
WeightsColumn = NULL,
TransformNumericColumns = NULL,
Methods = c("Asinh", "Asin", "Log", "LogPlus1", "Sqrt", "Logit"),
# Model args
GridTune = FALSE,
GridStrategy = "Cartesian",
StoppingRounds = 10,
MaxRunTimeSecs = 3600 * 24 * 7,
MaxModelsInGrid = 10,
Distribution = "gaussian",
Link = "identity",
TweedieLinkPower = NULL,
TweedieVariancePower = NULL,
RandomDistribution = NULL,
RandomLink = NULL,
Solver = "AUTO",
Alpha = NULL,
Lambda = NULL,
LambdaSearch = FALSE,
NLambdas = -1,
Standardize = TRUE,
RemoveCollinearColumns = FALSE,
InterceptInclude = TRUE,
NonNegativeCoefficients = FALSE)H2O-AutoML Example
# Create some dummy correlated data with numeric and categorical features
data <- AutoQuant::FakeDataGenerator(
Correlation = 0.85,
N = 1000,
ID = 2,
ZIP = 0,
AddDate = FALSE,
Classification = FALSE,
MultiClass = FALSE)
# Run function
TestModel <- AutoQuant::AutoH2oMLRegression(
# Compute management
MaxMem = "32G",
NThreads = max(1, parallel::detectCores()-2),
H2OShutdown = TRUE,
IfSaveModel = "mojo",
# Model evaluation
eval_metric = "RMSE",
NumOfParDepPlots = 3,
# Metadata arguments
model_path = NULL,
metadata_path = NULL,
ModelID = "FirstModel",
ReturnModelObjects = TRUE,
SaveModelObjects = FALSE,
# Data arguments
TrainOnFull = FALSE,
ValidationData = NULL,
TestData = NULL,
TargetColumnName = "Adrian",
FeatureColNames = names(data)[!names(data) %in% c("IDcol_1", "IDcol_2","Adrian")],
TransformNumericColumns = NULL,
Methods = c("Asinh", "Asin", "Log", "LogPlus1", "Logit"),
# Model args
GridTune = FALSE,
ExcludeAlgos = NULL,
Trees = 50,
MaxModelsInGrid = 10)H2O-GAM Example
# Create some dummy correlated data
data <- AutoQuant::FakeDataGenerator(
Correlation = 0.85,
N = 1000,
ID = 2,
ZIP = 0,
AddDate = FALSE,
Classification = FALSE,
MultiClass = FALSE)
# Define GAM Columns to use - up to 9 are allowed
GamCols <- names(which(unlist(lapply(data, is.numeric))))
GamCols <- GamCols[!GamCols %in% c("Adrian","IDcol_1","IDcol_2")]
GamCols <- GamCols[1L:(min(9L,length(GamCols)))]
# Run function
TestModel <- AutoQuant::AutoH2oGAMRegression(
# Compute management
MaxMem = {gc();paste0(as.character(floor(as.numeric(system("awk '/MemFree/ {print $2}' /proc/meminfo", intern=TRUE)) / 1000000)),"G")},
NThreads = max(1, parallel::detectCores()-2),
H2OShutdown = TRUE,
H2OStartUp = TRUE,
IfSaveModel = "mojo",
# Model evaluation:
eval_metric = "RMSE",
NumOfParDepPlots = 3,
# Metadata arguments:
model_path = NULL,
metadata_path = NULL,
ModelID = "FirstModel",
ReturnModelObjects = TRUE,
SaveModelObjects = FALSE,
SaveInfoToPDF = FALSE,
# Data arguments:
data = data,
TrainOnFull = FALSE,
ValidationData = NULL,
TestData = NULL,
TargetColumnName = "Adrian",
FeatureColNames = names(data)[!names(data) %in% c("IDcol_1", "IDcol_2","Adrian")],
InteractionColNumbers = NULL,
WeightsColumn = NULL,
GamColNames = GamCols,
TransformNumericColumns = NULL,
Methods = c("Asinh", "Asin", "Log", "LogPlus1", "Sqrt", "Logit"),
# Model args
num_knots = NULL,
keep_gam_cols = TRUE,
GridTune = FALSE,
GridStrategy = "Cartesian",
StoppingRounds = 10,
MaxRunTimeSecs = 3600 * 24 * 7,
MaxModelsInGrid = 10,
Distribution = "gaussian",
Link = "Family_Default",
TweedieLinkPower = NULL,
TweedieVariancePower = NULL,
Solver = "AUTO",
Alpha = NULL,
Lambda = NULL,
LambdaSearch = FALSE,
NLambdas = -1,
Standardize = TRUE,
RemoveCollinearColumns = FALSE,
InterceptInclude = TRUE,
NonNegativeCoefficients = FALSE)click to expand
Classification Description
The Auto_Classifier() models handle a multitude of tasks. In order:
- Convert your data to data.table format for faster processing
- Create train, validation, and test data if you didn't supply those directly to the function
- Consoldate columns that are used for modeling and what is to be kept for data returned
- Dichotomize categorical variables (for AutoXGBoostRegression) and save the factor levels for scoring in a way that guarentees consistency across training, validation, and test data sets
- Saves the final column names for modeling to a csv for later reference
- Handles the data conversion to the appropriate type, based on model type (CatBoost, H2O, and XGBoost)
- Multi-armed bandit hyperparameter tuning using randomized probability matching, if you choose to grid tune
- Build the grid tuned models
- Collect the evaluation metrics for each grid tune run
- Identify the best model of the set of models built in the grid tuning setup
- Save the hyperparameters from the winning grid tuned model
- Build the final model based on the best model from the grid tuning model search
- Collect evaluation metrics based on performance on test data
- Store the final predictions with the associated test data and other columns you want included in that set
- Build out and save an Evaluation Calibration Line Plot
- Build out and save an ROC plot with the top 5 models used in grid-tuning (includes the winning model)
- Generate and save Variable Importance data
- Generate and save Partital Dependence Calibration Line Plots
- Return all the objects generated in a named list for immediate use
CatBoost Example
# Create some dummy correlated data
data <- AutoQuant::FakeDataGenerator(
Correlation = 0.85,
N = 10000,
ID = 2,
ZIP = 0,
AddDate = FALSE,
Classification = TRUE,
MultiClass = FALSE)
# Run function
TestModel <- AutoQuant::AutoCatBoostClassifier(
# GPU or CPU and the number of available GPUs
task_type = 'GPU',
NumGPUs = 1,
TrainOnFull = FALSE,
DebugMode = FALSE,
# Metadata args
OutputSelection = c('Score_TrainData', 'Importance', 'EvalPlots', 'Metrics', 'PDF'),
ModelID = 'Test_Model_1',
model_path = normalizePath('./'),
metadata_path = normalizePath('./'),
SaveModelObjects = FALSE,
ReturnModelObjects = TRUE,
SaveInfoToPDF = FALSE,
# Data args
data = data,
ValidationData = NULL,
TestData = NULL,
TargetColumnName = 'Adrian',
FeatureColNames = names(data)[!names(data) %in%
c('IDcol_1','IDcol_2','Adrian')],
PrimaryDateColumn = NULL,
WeightsColumnName = NULL,
IDcols = c('IDcol_1','IDcol_2'),
# Evaluation args
ClassWeights = c(1L,1L),
CostMatrixWeights = c(1,0,0,1),
EvalMetric = 'AUC',
grid_eval_metric = 'MCC',
LossFunction = 'Logloss',
MetricPeriods = 10L,
NumOfParDepPlots = ncol(data)-1L-2L,
# Grid tuning args
PassInGrid = NULL,
GridTune = FALSE,
MaxModelsInGrid = 30L,
MaxRunsWithoutNewWinner = 20L,
MaxRunMinutes = 24L*60L,
BaselineComparison = 'default',
# ML args
Trees = 1000,
Depth = 9,
LearningRate = NULL,
L2_Leaf_Reg = NULL,
model_size_reg = 0.5,
langevin = FALSE,
diffusion_temperature = 10000,
RandomStrength = 1,
BorderCount = 128,
RSM = 1,
BootStrapType = 'Bayesian',
GrowPolicy = 'SymmetricTree',
feature_border_type = 'GreedyLogSum',
sampling_unit = 'Object',
subsample = NULL,
score_function = 'Cosine',
min_data_in_leaf = 1)XGBoost Example
# Create some dummy correlated data
data <- AutoQuant::FakeDataGenerator(
Correlation = 0.85,
N = 1000L,
ID = 2L,
ZIP = 0L,
AddDate = FALSE,
Classification = TRUE,
MultiClass = FALSE)
# Run function
TestModel <- AutoQuant::AutoXGBoostClassifier(
# GPU or CPU
TreeMethod = "hist",
NThreads = parallel::detectCores(),
# Metadata args
OutputSelection = c("Importances", "EvalPlots", "EvalMetrics", "PDFs", "Score_TrainData"),
model_path = normalizePath("./"),
metadata_path = NULL,
ModelID = "Test_Model_1",
EncodingMethod = "binary",
ReturnFactorLevels = TRUE,
ReturnModelObjects = TRUE,
SaveModelObjects = FALSE,
SaveInfoToPDF = FALSE,
# Data args
data = data,
TrainOnFull = FALSE,
ValidationData = NULL,
TestData = NULL,
TargetColumnName = "Adrian",
FeatureColNames = names(data)[!names(data) %in%
c("IDcol_1", "IDcol_2","Adrian")],
WeightsColumnName = NULL,
IDcols = c("IDcol_1","IDcol_2"),
# Model evaluation
LossFunction = 'reg:logistic',
CostMatrixWeights = c(1,0,0,1),
eval_metric = "auc",
grid_eval_metric = "MCC",
NumOfParDepPlots = 3L,
# Grid tuning args
PassInGrid = NULL,
GridTune = FALSE,
BaselineComparison = "default",
MaxModelsInGrid = 10L,
MaxRunsWithoutNewWinner = 20L,
MaxRunMinutes = 24L*60L,
Verbose = 1L,
# ML args
Trees = 500L,
eta = 0.30,
max_depth = 9L,
min_child_weight = 1.0,
subsample = 1,
colsample_bytree = 1,
DebugMode = FALSE)LightGBM Example
# Create some dummy correlated data
data <- AutoQuant::FakeDataGenerator(
Correlation = 0.85,
N = 1000,
ID = 2,
ZIP = 0,
AddDate = FALSE,
Classification = FALSE,
MultiClass = FALSE)
# Run function
TestModel <- AutoQuant::AutoLightGBMClassifier(
# Metadata args
OutputSelection = c("Importances","EvalPlots","EvalMetrics","Score_TrainData"),
model_path = normalizePath("./"),
metadata_path = NULL,
ModelID = "Test_Model_1",
NumOfParDepPlots = 3L,
EncodingMethod = "credibility",
ReturnFactorLevels = TRUE,
ReturnModelObjects = TRUE,
SaveModelObjects = FALSE,
SaveInfoToPDF = FALSE,
DebugMode = FALSE,
# Data args
data = data,
TrainOnFull = FALSE,
ValidationData = NULL,
TestData = NULL,
TargetColumnName = "Adrian",
FeatureColNames = names(data)[!names(data) %in% c("IDcol_1", "IDcol_2","Adrian")],
PrimaryDateColumn = NULL,
WeightsColumnName = NULL,
IDcols = c("IDcol_1","IDcol_2"),
# Grid parameters
GridTune = FALSE,
grid_eval_metric = 'Utility',
BaselineComparison = 'default',
MaxModelsInGrid = 10L,
MaxRunsWithoutNewWinner = 20L,
MaxRunMinutes = 24L*60L,
PassInGrid = NULL,
# Core parameters
# https://lightgbm.readthedocs.io/en/latest/Parameters.html#core-parameters
input_model = NULL, # continue training a model that is stored to file
task = "train",
device_type = 'CPU',
NThreads = parallel::detectCores() / 2,
objective = 'binary',
metric = 'binary_logloss',
boosting = 'gbdt',
LinearTree = FALSE,
Trees = 50L,
eta = NULL,
num_leaves = 31,
deterministic = TRUE,
# Learning Parameters
# https://lightgbm.readthedocs.io/en/latest/Parameters.html#learning-control-parameters
force_col_wise = FALSE,
force_row_wise = FALSE,
max_depth = NULL,
min_data_in_leaf = 20,
min_sum_hessian_in_leaf = 0.001,
bagging_freq = 0,
bagging_fraction = 1.0,
feature_fraction = 1.0,
feature_fraction_bynode = 1.0,
extra_trees = FALSE,
early_stopping_round = 10,
first_metric_only = TRUE,
max_delta_step = 0.0,
lambda_l1 = 0.0,
lambda_l2 = 0.0,
linear_lambda = 0.0,
min_gain_to_split = 0,
drop_rate_dart = 0.10,
max_drop_dart = 50,
skip_drop_dart = 0.50,
uniform_drop_dart = FALSE,
top_rate_goss = FALSE,
other_rate_goss = FALSE,
monotone_constraints = NULL,
monotone_constraints_method = "advanced",
monotone_penalty = 0.0,
forcedsplits_filename = NULL, # use for AutoStack option; .json file
refit_decay_rate = 0.90,
path_smooth = 0.0,
# IO Dataset Parameters
# https://lightgbm.readthedocs.io/en/latest/Parameters.html#io-parameters
max_bin = 255,
min_data_in_bin = 3,
data_random_seed = 1,
is_enable_sparse = TRUE,
enable_bundle = TRUE,
use_missing = TRUE,
zero_as_missing = FALSE,
two_round = FALSE,
# Convert Parameters
convert_model = NULL,
convert_model_language = "cpp",
# Objective Parameters
# https://lightgbm.readthedocs.io/en/latest/Parameters.html#objective-parameters
boost_from_average = TRUE,
is_unbalance = FALSE,
scale_pos_weight = 1.0,
# Metric Parameters (metric is in Core)
# https://lightgbm.readthedocs.io/en/latest/Parameters.html#metric-parameters
is_provide_training_metric = TRUE,
eval_at = c(1,2,3,4,5),
# Network Parameters
# https://lightgbm.readthedocs.io/en/latest/Parameters.html#network-parameters
num_machines = 1,
# GPU Parameters
# https://lightgbm.readthedocs.io/en/latest/Parameters.html#gpu-parameters
gpu_platform_id = -1,
gpu_device_id = -1,
gpu_use_dp = TRUE,
num_gpu = 1)H2O-GBM Example
# Create some dummy correlated data
data <- AutoQuant::FakeDataGenerator(
Correlation = 0.85,
N = 1000L,
ID = 2L,
ZIP = 0L,
AddDate = FALSE,
Classification = TRUE,
MultiClass = FALSE)
TestModel <- AutoQuant::AutoH2oGBMClassifier(
# Compute management
MaxMem = {gc();paste0(as.character(floor(as.numeric(system("awk '/MemFree/ {print $2}' /proc/meminfo", intern=TRUE)) / 1000000)),"G")},
NThreads = max(1, parallel::detectCores()-2),
H2OShutdown = TRUE,
H2OStartUp = TRUE,
IfSaveModel = "mojo",
# Model evaluation
NumOfParDepPlots = 3,
# Metadata arguments:
model_path = normalizePath("./"),
metadata_path = file.path(normalizePath("./")),
ModelID = "FirstModel",
ReturnModelObjects = TRUE,
SaveModelObjects = FALSE,
SaveInfoToPDF = FALSE,
# Data arguments
data = data,
TrainOnFull = FALSE,
ValidationData = NULL,
TestData = NULL,
TargetColumnName = "Adrian",
FeatureColNames = names(data)[!names(data) %in% c("IDcol_1", "IDcol_2","Adrian")],
WeightsColumn = NULL,
# ML grid tuning args
GridTune = FALSE,
GridStrategy = "Cartesian",
MaxRuntimeSecs = 60*60*24,
StoppingRounds = 10,
MaxModelsInGrid = 2,
# Model args
Trees = 50,
LearnRate = 0.10,
LearnRateAnnealing = 1,
eval_metric = "auc",
Distribution = "bernoulli",
MaxDepth = 20,
SampleRate = 0.632,
ColSampleRate = 1,
ColSampleRatePerTree = 1,
ColSampleRatePerTreeLevel = 1,
MinRows = 1,
NBins = 20,
NBinsCats = 1024,
NBinsTopLevel = 1024,
HistogramType = "AUTO",
CategoricalEncoding = "AUTO")H2O-DRF Example
# Create some dummy correlated data
data <- AutoQuant::FakeDataGenerator(
Correlation = 0.85,
N = 1000L,
ID = 2L,
ZIP = 0L,
AddDate = FALSE,
Classification = TRUE,
MultiClass = FALSE)
TestModel <- AutoQuant::AutoH2oDRFClassifier(
# Compute management
MaxMem = {gc();paste0(as.character(floor(as.numeric(system("awk '/MemFree/ {print $2}' /proc/meminfo", intern=TRUE)) / 1000000)),"G")},
NThreads = max(1L, parallel::detectCores() - 2L),
IfSaveModel = "mojo",
H2OShutdown = FALSE,
H2OStartUp = TRUE,
# Metadata arguments:
eval_metric = "auc",
NumOfParDepPlots = 3L,
# Data arguments:
model_path = normalizePath("./"),
metadata_path = NULL,
ModelID = "FirstModel",
ReturnModelObjects = TRUE,
SaveModelObjects = FALSE,
SaveInfoToPDF = FALSE,
# Model evaluation:
data,
TrainOnFull = FALSE,
ValidationData = NULL,
TestData = NULL,
TargetColumnName = "Adrian",
FeatureColNames = names(data)[!names(data) %in% c("IDcol_1", "IDcol_2", "Adrian")],
WeightsColumn = NULL,
# Grid Tuning Args
GridStrategy = "Cartesian",
GridTune = FALSE,
MaxModelsInGrid = 10,
MaxRuntimeSecs = 60*60*24,
StoppingRounds = 10,
# Model args
Trees = 50L,
MaxDepth = 20,
SampleRate = 0.632,
MTries = -1,
ColSampleRatePerTree = 1,
ColSampleRatePerTreeLevel = 1,
MinRows = 1,
NBins = 20,
NBinsCats = 1024,
NBinsTopLevel = 1024,
HistogramType = "AUTO",
CategoricalEncoding = "AUTO")H2O-GLM Example
# Create some dummy correlated data with numeric and categorical features
data <- AutoQuant::FakeDataGenerator(
Correlation = 0.85,
N = 1000L,
ID = 2L,
ZIP = 0L,
AddDate = FALSE,
Classification = TRUE,
MultiClass = FALSE)
# Run function
TestModel <- AutoQuant::AutoH2oGLMClassifier(
# Compute management
MaxMem = {gc();paste0(as.character(floor(as.numeric(system("awk '/MemFree/ {print $2}' /proc/meminfo", intern=TRUE)) / 1000000)),"G")},
NThreads = max(1, parallel::detectCores()-2),
H2OShutdown = TRUE,
H2OStartUp = TRUE,
IfSaveModel = "mojo",
# Model evaluation args
eval_metric = "auc",
NumOfParDepPlots = 3,
# Metadata args
model_path = NULL,
metadata_path = NULL,
ModelID = "FirstModel",
ReturnModelObjects = TRUE,
SaveModelObjects = FALSE,
SaveInfoToPDF = FALSE,
# Data args
data = data,
TrainOnFull = FALSE,
ValidationData = NULL,
TestData = NULL,
TargetColumnName = "Adrian",
FeatureColNames = names(data)[!names(data) %in%
c("IDcol_1", "IDcol_2","Adrian")],
RandomColNumbers = NULL,
InteractionColNumbers = NULL,
WeightsColumn = NULL,
# ML args
GridTune = FALSE,
GridStrategy = "Cartesian",
StoppingRounds = 10,
MaxRunTimeSecs = 3600 * 24 * 7,
MaxModelsInGrid = 10,
Distribution = "binomial",
Link = "logit",
RandomDistribution = NULL,
RandomLink = NULL,
Solver = "AUTO",
Alpha = NULL,
Lambda = NULL,
LambdaSearch = FALSE,
NLambdas = -1,
Standardize = TRUE,
RemoveCollinearColumns = FALSE,
InterceptInclude = TRUE,
NonNegativeCoefficients = FALSE)H2O-AutoML Example
# Create some dummy correlated data with numeric and categorical features
data <- AutoQuant::FakeDataGenerator(
Correlation = 0.85,
N = 1000L,
ID = 2L,
ZIP = 0L,
AddDate = FALSE,
Classification = TRUE,
MultiClass = FALSE)
TestModel <- AutoQuant::AutoH2oMLClassifier(
data,
TrainOnFull = FALSE,
ValidationData = NULL,
TestData = NULL,
TargetColumnName = "Adrian",
FeatureColNames = names(data)[!names(data) %in% c("IDcol_1", "IDcol_2","Adrian")],
ExcludeAlgos = NULL,
eval_metric = "auc",
Trees = 50,
MaxMem = "32G",
NThreads = max(1, parallel::detectCores()-2),
MaxModelsInGrid = 10,
model_path = normalizePath("./"),
metadata_path = file.path(normalizePath("./"), "MetaData"),
ModelID = "FirstModel",
NumOfParDepPlots = 3,
ReturnModelObjects = TRUE,
SaveModelObjects = FALSE,
IfSaveModel = "mojo",
H2OShutdown = FALSE,
HurdleModel = FALSE)H2O-GAM Example
# Create some dummy correlated data
data <- AutoQuant::FakeDataGenerator(
Correlation = 0.85,
N = 1000,
ID = 2,
ZIP = 0,
AddDate = FALSE,
Classification = TRUE,
MultiClass = FALSE)
# Define GAM Columns to use - up to 9 are allowed
GamCols <- names(which(unlist(lapply(data, is.numeric))))
GamCols <- GamCols[!GamCols %in% c("Adrian","IDcol_1","IDcol_2")]
GamCols <- GamCols[1L:(min(9L,length(GamCols)))]
# Run function
TestModel <- AutoQuant::AutoH2oGAMClassifier(
# Compute management
MaxMem = {gc();paste0(as.character(floor(as.numeric(system("awk '/MemFree/ {print $2}' /proc/meminfo", intern=TRUE)) / 1000000)),"G")},
NThreads = max(1, parallel::detectCores()-2),
H2OShutdown = TRUE,
H2OStartUp = TRUE,
IfSaveModel = "mojo",
# Model evaluation:
eval_metric = "auc",
NumOfParDepPlots = 3,
# Metadata arguments:
model_path = NULL,
metadata_path = NULL,
ModelID = "FirstModel",
ReturnModelObjects = TRUE,
SaveModelObjects = FALSE,
SaveInfoToPDF = FALSE,
# Data arguments:
data = data,
TrainOnFull = FALSE,
ValidationData = NULL,
TestData = NULL,
TargetColumnName = "Adrian",
FeatureColNames = names(data)[!names(data) %in% c("IDcol_1", "IDcol_2","Adrian")],
WeightsColumn = NULL,
GamColNames = GamCols,
# ML args
num_knots = NULL,
keep_gam_cols = TRUE,
GridTune = FALSE,
GridStrategy = "Cartesian",
StoppingRounds = 10,
MaxRunTimeSecs = 3600 * 24 * 7,
MaxModelsInGrid = 10,
Distribution = "binomial",
Link = "logit",
Solver = "AUTO",
Alpha = NULL,
Lambda = NULL,
LambdaSearch = FALSE,
NLambdas = -1,
Standardize = TRUE,
RemoveCollinearColumns = FALSE,
InterceptInclude = TRUE,
NonNegativeCoefficients = FALSE)click to expand
MultiClass Description
The Auto_MultiClass() models handle a multitude of tasks. In order:
- Convert your data to data.table format for faster processing
- Create train, validation, and test data if you didn't supply those directly to the function
- Consoldate columns that are used for modeling and what is to be kept for data returned
- Dichotomize categorical variables (for AutoXGBoostRegression) and save the factor levels for scoring in a way that guarentees consistency across training, validation, and test data sets
- Saves the final column names for modeling to a csv for later reference
- Ensures the target levels are consistent across train, validate, and test sets and save the levels to file
- Handles the data conversion to the appropriate type, based on model type (CatBoost, H2O, and XGBoost)
- Multi-armed bandit hyperparameter tuning using randomized probability matching, if you choose to grid tune
- Build the grid tuned models
- Collect the evaluation metrics for each grid tune run
- Identify the best model of the set of models built in the grid tuning setup
- Save the hyperparameters from the winning grid tuned model
- Build the final model based on the best model from the grid tuning model search
- Collect evaluation metrics based on performance on test data
- Store the final predictions with the associated test data and other columns you want included in that set
- Generate and save Variable Importance data
- Return all the objects generated in a named list for immediate use
CatBoost Example
# Create some dummy correlated data
data <- AutoQuant::FakeDataGenerator(
Correlation = 0.85,
N = 10000L,
ID = 2L,
ZIP = 0L,
AddDate = FALSE,
Classification = FALSE,
MultiClass = TRUE)
# Run function
TestModel <- AutoQuant::AutoCatBoostMultiClass(
# GPU or CPU and the number of available GPUs
task_type = 'GPU',
NumGPUs = 1,
TrainOnFull = FALSE,
DebugMode = FALSE,
# Metadata args
OutputSelection = c('Importances', 'EvalPlots', 'EvalMetrics', 'Score_TrainData'),
ModelID = 'Test_Model_1',
model_path = normalizePath('./'),
metadata_path = normalizePath('./'),
SaveModelObjects = FALSE,
ReturnModelObjects = TRUE,
# Data args
data = data,
ValidationData = NULL,
TestData = NULL,
TargetColumnName = 'Adrian',
FeatureColNames = names(data)[!names(data) %in%
c('IDcol_1', 'IDcol_2','Adrian')],
PrimaryDateColumn = NULL,
WeightsColumnName = NULL,
ClassWeights = c(1L,1L,1L,1L,1L),
IDcols = c('IDcol_1','IDcol_2'),
# Model evaluation
eval_metric = 'MCC',
loss_function = 'MultiClassOneVsAll',
grid_eval_metric = 'Accuracy',
MetricPeriods = 10L,
NumOfParDepPlots = 3,
# Grid tuning args
PassInGrid = NULL,
GridTune = TRUE,
MaxModelsInGrid = 30L,
MaxRunsWithoutNewWinner = 20L,
MaxRunMinutes = 24L*60L,
BaselineComparison = 'default',
# ML args
langevin = FALSE,
diffusion_temperature = 10000,
Trees = seq(100L, 500L, 50L),
Depth = seq(4L, 8L, 1L),
LearningRate = seq(0.01,0.10,0.01),
L2_Leaf_Reg = seq(1.0, 10.0, 1.0),
RandomStrength = 1,
BorderCount = 254,
RSM = c(0.80, 0.85, 0.90, 0.95, 1.0),
BootStrapType = c('Bayesian', 'Bernoulli', 'Poisson', 'MVS', 'No'),
GrowPolicy = c('SymmetricTree', 'Depthwise', 'Lossguide'),
model_size_reg = 0.5,
feature_border_type = 'GreedyLogSum',
sampling_unit = 'Object',
subsample = NULL,
score_function = 'Cosine',
min_data_in_leaf = 1)XGBoost Example
# Create some dummy correlated data
data <- AutoQuant::FakeDataGenerator(
Correlation = 0.85,
N = 1000L,
ID = 2L,
ZIP = 0L,
AddDate = FALSE,
Classification = FALSE,
MultiClass = TRUE)
# Run function
TestModel <- AutoQuant::AutoXGBoostMultiClass(
# GPU or CPU
TreeMethod = "hist",
NThreads = parallel::detectCores(),
# Metadata args
OutputSelection = c("Importances", "EvalPlots", "EvalMetrics", "PDFs", "Score_TrainData"),
model_path = normalizePath("./"),
metadata_path = normalizePath("./"),
ModelID = "Test_Model_1",
EncodingMethod = "binary",
ReturnFactorLevels = TRUE,
ReturnModelObjects = TRUE,
SaveModelObjects = FALSE,
# Data args
data = data,
TrainOnFull = FALSE,
ValidationData = NULL,
TestData = NULL,
TargetColumnName = "Adrian",
FeatureColNames = names(data)[!names(data) %in%
c("IDcol_1", "IDcol_2","Adrian")],
WeightsColumnName = NULL,
IDcols = c("IDcol_1","IDcol_2"),
# Model evaluation args
eval_metric = "merror",
LossFunction = 'multi:softprob',
grid_eval_metric = "accuracy",
NumOfParDepPlots = 3L,
# Grid tuning args
PassInGrid = NULL,
GridTune = FALSE,
BaselineComparison = "default",
MaxModelsInGrid = 10L,
MaxRunsWithoutNewWinner = 20L,
MaxRunMinutes = 24L*60L,
Verbose = 1L,
DebugMode = FALSE,
# ML args
Trees = 50L,
eta = 0.05,
max_depth = 4L,
min_child_weight = 1.0,
subsample = 0.55,
colsample_bytree = 0.55)LightGBM Example
# Create some dummy correlated data
data <- AutoQuant::FakeDataGenerator(
Correlation = 0.85,
N = 1000,
ID = 2,
ZIP = 0,
AddDate = FALSE,
Classification = FALSE,
MultiClass = FALSE)
# Run function
TestModel <- AutoQuant::AutoLightGBMMultiClass(
# Metadata args
OutputSelection = c("Importances","EvalPlots","EvalMetrics","Score_TrainData"),
model_path = normalizePath("./"),
metadata_path = NULL,
ModelID = "Test_Model_1",
NumOfParDepPlots = 3L,
EncodingMethod = "credibility",
ReturnFactorLevels = TRUE,
ReturnModelObjects = TRUE,
SaveModelObjects = FALSE,
SaveInfoToPDF = FALSE,
DebugMode = FALSE,
# Data args
data = data,
TrainOnFull = FALSE,
ValidationData = NULL,
TestData = NULL,
TargetColumnName = "Adrian",
FeatureColNames = names(data)[!names(data) %in% c("IDcol_1", "IDcol_2","Adrian")],
PrimaryDateColumn = NULL,
WeightsColumnName = NULL,
IDcols = c("IDcol_1","IDcol_2"),
# Grid parameters
GridTune = FALSE,
grid_eval_metric = 'microauc',
BaselineComparison = 'default',
MaxModelsInGrid = 10L,
MaxRunsWithoutNewWinner = 20L,
MaxRunMinutes = 24L*60L,
PassInGrid = NULL,
# Core parameters
# https://lightgbm.readthedocs.io/en/latest/Parameters.html#core-parameters
input_model = NULL, # continue training a model that is stored to file
task = "train",
device_type = 'CPU',
NThreads = parallel::detectCores() / 2,
objective = 'multiclass',
multi_error_top_k = 1,
metric = 'multi_logloss',
boosting = 'gbdt',
LinearTree = FALSE,
Trees = 50L,
eta = NULL,
num_leaves = 31,
deterministic = TRUE,
# Learning Parameters
# https://lightgbm.readthedocs.io/en/latest/Parameters.html#learning-control-parameters
force_col_wise = FALSE,
force_row_wise = FALSE,
max_depth = NULL,
min_data_in_leaf = 20,
min_sum_hessian_in_leaf = 0.001,
bagging_freq = 0,
bagging_fraction = 1.0,
feature_fraction = 1.0,
feature_fraction_bynode = 1.0,
extra_trees = FALSE,
early_stopping_round = 10,
first_metric_only = TRUE,
max_delta_step = 0.0,
lambda_l1 = 0.0,
lambda_l2 = 0.0,
linear_lambda = 0.0,
min_gain_to_split = 0,
drop_rate_dart = 0.10,
max_drop_dart = 50,
skip_drop_dart = 0.50,
uniform_drop_dart = FALSE,
top_rate_goss = FALSE,
other_rate_goss = FALSE,
monotone_constraints = NULL,
monotone_constraints_method = "advanced",
monotone_penalty = 0.0,
forcedsplits_filename = NULL, # use for AutoStack option; .json file
refit_decay_rate = 0.90,
path_smooth = 0.0,
# IO Dataset Parameters
# https://lightgbm.readthedocs.io/en/latest/Parameters.html#io-parameters
max_bin = 255,
min_data_in_bin = 3,
data_random_seed = 1,
is_enable_sparse = TRUE,
enable_bundle = TRUE,
use_missing = TRUE,
zero_as_missing = FALSE,
two_round = FALSE,
# Convert Parameters
convert_model = NULL,
convert_model_language = "cpp",
# Objective Parameters
# https://lightgbm.readthedocs.io/en/latest/Parameters.html#objective-parameters
boost_from_average = TRUE,
is_unbalance = FALSE,
scale_pos_weight = 1.0,
# Metric Parameters (metric is in Core)
# https://lightgbm.readthedocs.io/en/latest/Parameters.html#metric-parameters
is_provide_training_metric = TRUE,
eval_at = c(1,2,3,4,5),
# Network Parameters
# https://lightgbm.readthedocs.io/en/latest/Parameters.html#network-parameters
num_machines = 1,
# GPU Parameters
# https://lightgbm.readthedocs.io/en/latest/Parameters.html#gpu-parameters
gpu_platform_id = -1,
gpu_device_id = -1,
gpu_use_dp = TRUE,
num_gpu = 1)H2O-GBM Example
# Create some dummy correlated data
data <- AutoQuant::FakeDataGenerator(
Correlation = 0.85,
N = 1000,
ID = 2,
ZIP = 0,
AddDate = FALSE,
Classification = FALSE,
MultiClass = TRUE)
# Run function
TestModel <- AutoQuant::AutoH2oGBMMultiClass(
data,
TrainOnFull = FALSE,
ValidationData = NULL,
TestData = NULL,
TargetColumnName = "Adrian",
FeatureColNames = names(data)[!names(data) %in% c("IDcol_1", "IDcol_2","Adrian")],
WeightsColumn = NULL,
eval_metric = "logloss",
MaxMem = {gc();paste0(as.character(floor(as.numeric(system("awk '/MemFree/ {print $2}' /proc/meminfo", intern=TRUE)) / 1000000)),"G")},
NThreads = max(1, parallel::detectCores()-2),
model_path = normalizePath("./"),
metadata_path = file.path(normalizePath("./")),
ModelID = "FirstModel",
ReturnModelObjects = TRUE,
SaveModelObjects = FALSE,
IfSaveModel = "mojo",
H2OShutdown = TRUE,
H2OStartUp = TRUE,
# Model args
GridTune = FALSE,
GridStrategy = "Cartesian",
MaxRuntimeSecs = 60*60*24,
StoppingRounds = 10,
MaxModelsInGrid = 2,
Trees = 50,
LearnRate = 0.10,
LearnRateAnnealing = 1,
eval_metric = "RMSE",
Distribution = "multinomial",
MaxDepth = 20,
SampleRate = 0.632,
ColSampleRate = 1,
ColSampleRatePerTree = 1,
ColSampleRatePerTreeLevel = 1,
MinRows = 1,
NBins = 20,
NBinsCats = 1024,
NBinsTopLevel = 1024,
HistogramType = "AUTO",
CategoricalEncoding = "AUTO")H2O-DRF Example
# Create some dummy correlated data
data <- AutoQuant::FakeDataGenerator(
Correlation = 0.85,
N = 1000L,
ID = 2L,
ZIP = 0L,
AddDate = FALSE,
Classification = FALSE,
MultiClass = TRUE)
# Run function
TestModel <- AutoQuant::AutoH2oDRFMultiClass(
data,
TrainOnFull = FALSE,
ValidationData = NULL,
TestData = NULL,
TargetColumnName = "Adrian",
FeatureColNames = names(data)[!names(data) %in% c("IDcol_1", "IDcol_2","Adrian")],
WeightsColumn = NULL,
eval_metric = "logloss",
MaxMem = {gc();paste0(as.character(floor(as.numeric(system("awk '/MemFree/ {print $2}' /proc/meminfo", intern=TRUE)) / 1000000)),"G")},
NThreads = max(1, parallel::detectCores()-2),
model_path = normalizePath("./"),
metadata_path = file.path(normalizePath("./")),
ModelID = "FirstModel",
ReturnModelObjects = TRUE,
SaveModelObjects = FALSE,
IfSaveModel = "mojo",
H2OShutdown = FALSE,
H2OStartUp = TRUE,
# Grid Tuning Args
GridStrategy = "Cartesian",
GridTune = FALSE,
MaxModelsInGrid = 10,
MaxRuntimeSecs = 60*60*24,
StoppingRounds = 10,
# ML args
Trees = 50,
MaxDepth = 20,
SampleRate = 0.632,
MTries = -1,
ColSampleRatePerTree = 1,
ColSampleRatePerTreeLevel = 1,
MinRows = 1,
NBins = 20,
NBinsCats = 1024,
NBinsTopLevel = 1024,
HistogramType = "AUTO",
CategoricalEncoding = "AUTO")H2O-GLM Example
# Create some dummy correlated data with numeric and categorical features
data <- AutoQuant::FakeDataGenerator(
Correlation = 0.85,
N = 1000L,
ID = 2L,
ZIP = 0L,
AddDate = FALSE,
Classification = FALSE,
MultiClass = TRUE)
# Run function
TestModel <- AutoQuant::AutoH2oGLMMultiClass(
# Compute management
MaxMem = {gc();paste0(as.character(floor(as.numeric(system("awk '/MemFree/ {print $2}' /proc/meminfo", intern=TRUE)) / 1000000)),"G")},
NThreads = max(1, parallel::detectCores()-2),
H2OShutdown = TRUE,
H2OStartUp = TRUE,
IfSaveModel = "mojo",
# Model evaluation:
eval_metric = "logloss",
NumOfParDepPlots = 3,
# Metadata arguments:
model_path = NULL,
metadata_path = NULL,
ModelID = "FirstModel",
ReturnModelObjects = TRUE,
SaveModelObjects = FALSE,
SaveInfoToPDF = FALSE,
# Data arguments:
data = data,
TrainOnFull = FALSE,
ValidationData = NULL,
TestData = NULL,
TargetColumnName = "Adrian",
FeatureColNames = names(data)[!names(data) %in% c("IDcol_1", "IDcol_2","Adrian")],
RandomColNumbers = NULL,
InteractionColNumbers = NULL,
WeightsColumn = NULL,
# Model args
GridTune = FALSE,
GridStrategy = "Cartesian",
StoppingRounds = 10,
MaxRunTimeSecs = 3600 * 24 * 7,
MaxModelsInGrid = 10,
Distribution = "multinomial",
Link = "family_default",
RandomDistribution = NULL,
RandomLink = NULL,
Solver = "AUTO",
Alpha = NULL,
Lambda = NULL,
LambdaSearch = FALSE,
NLambdas = -1,
Standardize = TRUE,
RemoveCollinearColumns = FALSE,
InterceptInclude = TRUE,
NonNegativeCoefficients = FALSE)H2O-AutoML Example
# Create some dummy correlated data with numeric and categorical features
data <- AutoQuant::FakeDataGenerator(Correlation = 0.85, N = 1000, ID = 2, ZIP = 0, AddDate = FALSE, Classification = FALSE, MultiClass = TRUE)
# Run function
TestModel <- AutoQuant::AutoH2oMLMultiClass(
data,
TrainOnFull = FALSE,
ValidationData = NULL,
TestData = NULL,
TargetColumnName = "Adrian",
FeatureColNames = names(data)[!names(data) %in% c("IDcol_1", "IDcol_2","Adrian")],
ExcludeAlgos = NULL,
eval_metric = "logloss",
Trees = 50,
MaxMem = "32G",
NThreads = max(1, parallel::detectCores()-2),
MaxModelsInGrid = 10,
model_path = normalizePath("./"),
metadata_path = file.path(normalizePath("./"), "MetaData"),
ModelID = "FirstModel",
ReturnModelObjects = TRUE,
SaveModelObjects = FALSE,
IfSaveModel = "mojo",
H2OShutdown = FALSE,
HurdleModel = FALSE)H2O-GAM Example
# Create some dummy correlated data with numeric and categorical features
data <- AutoQuant::FakeDataGenerator(
Correlation = 0.85,
N = 1000L,
ID = 2L,
ZIP = 0L,
AddDate = FALSE,
Classification = FALSE,
MultiClass = TRUE)
# Define GAM Columns to use - up to 9 are allowed
GamCols <- names(which(unlist(lapply(data, is.numeric))))
GamCols <- GamCols[!GamCols %in% c("Adrian","IDcol_1","IDcol_2")]
GamCols <- GamCols[1L:(min(9L,length(GamCols)))]
# Run function
TestModel <- AutoQuant::AutoH2oGAMMultiClass(
data,
TrainOnFull = FALSE,
ValidationData = NULL,
TestData = NULL,
TargetColumnName = "Adrian",
FeatureColNames = names(data)[!names(data) %in% c("IDcol_1", "IDcol_2","Adrian")],
WeightsColumn = NULL,
GamColNames = GamCols,
eval_metric = "logloss",
MaxMem = {gc();paste0(as.character(floor(as.numeric(system("awk '/MemFree/ {print $2}' /proc/meminfo", intern=TRUE)) / 1000000)),"G")},
NThreads = max(1, parallel::detectCores()-2),
model_path = normalizePath("./"),
metadata_path = NULL,
ModelID = "FirstModel",
ReturnModelObjects = TRUE,
SaveModelObjects = FALSE,
IfSaveModel = "mojo",
H2OShutdown = FALSE,
H2OStartUp = TRUE,
# ML args
num_knots = NULL,
keep_gam_cols = TRUE,
GridTune = FALSE,
GridStrategy = "Cartesian",
StoppingRounds = 10,
MaxRunTimeSecs = 3600 * 24 * 7,
MaxModelsInGrid = 10,
Distribution = "multinomial",
Link = "Family_Default",
Solver = "AUTO",
Alpha = NULL,
Lambda = NULL,
LambdaSearch = FALSE,
NLambdas = -1,
Standardize = TRUE,
RemoveCollinearColumns = FALSE,
InterceptInclude = TRUE,
NonNegativeCoefficients = FALSE)Expand to view content
Scoring Description
AutoCatBoostScoring() is an automated scoring function that compliments the AutoCatBoost__() model training functions. This function requires you to supply features for scoring. It will run ModelDataPrep() to prepare your features for catboost data conversion and scoring. It will also handle and transformations and back-transformations if you utilized that feature in the regression training case.
AutoXGBoostScoring() is an automated scoring function that compliments the AutoXGBoost__() model training functions. This function requires you to supply features for scoring. It will run ModelDataPrep() and the CategoricalEncoding() functions to prepare your features for xgboost data conversion and scoring. It will also handle and transformations and back-transformations if you utilized that feature in the regression training case.
AutoLightGBMScoring() is an automated scoring function that compliments the AutoLightGBM__() model training functions. This function requires you to supply features for scoring. It will run ModelDataPrep() and the CategoricalEncoding() functions to prepare your features for lightgbm data conversion and scoring. It will also handle and transformations and back-transformations if you utilized that feature in the regression training case.
AutoH2OMLScoring() is an automated scoring function that compliments the AutoH2oGBM__() and AutoH2oDRF__() model training functions. This function requires you to supply features for scoring. It will run ModelDataPrep()to prepare your features for H2O data conversion and scoring. It will also handle transformations and back-transformations if you utilized that feature in the regression training case and didn't do it yourself before hand.
AutoCatBoostHurdleModelScoring() for scoring models developed with AutoCatBoostHurdleModel()
AutoLightGBMHurdleModelScoring() for scoring models developed with AutoLightGBMHurdleModel()
AutoXGBoostHurdleModelScoring() for scoring models developed with AutoXGBoostHurdleModel()
AutoCatBoost__() Examples
AutoCatBoostRegression() Scoring Example
# Create some dummy correlated data
data <- AutoQuant::FakeDataGenerator(
Correlation = 0.85,
N = 10000,
ID = 2,
ZIP = 0,
AddDate = FALSE,
Classification = FALSE,
MultiClass = FALSE)
# Copy data
data1 <- data.table::copy(data)
# Feature Colnames
Features <- names(data1)[!names(data1) %in% c("IDcol_1", "IDcol_2","DateTime","Adrian")]
# Run function
TestModel <- AutoQuant::AutoCatBoostRegression(
# GPU or CPU and the number of available GPUs
TrainOnFull = FALSE,
task_type = 'CPU',
NumGPUs = 1,
DebugMode = FALSE,
# Metadata args
OutputSelection = c('Importances','EvalPlots','EvalMetrics','Score_TrainData'),
ModelID = 'Test_Model_1',
model_path = getwd(),
metadata_path = getwd(),
SaveModelObjects = FALSE,
SaveInfoToPDF = FALSE,
ReturnModelObjects = TRUE,
# Data args
data = data1,
ValidationData = NULL,
TestData = NULL,
TargetColumnName = 'Adrian',
FeatureColNames = Features,
PrimaryDateColumn = NULL,
WeightsColumnName = NULL,
IDcols = c('IDcol_1','IDcol_2'),
TransformNumericColumns = 'Adrian',
Methods = c('Asinh','Asin','Log','LogPlus1','Sqrt','Logit'),
# Model evaluation
eval_metric = 'RMSE',
eval_metric_value = 1.5,
loss_function = 'RMSE',
loss_function_value = 1.5,
MetricPeriods = 10L,
NumOfParDepPlots = ncol(data1)-1L-2L,
# Grid tuning args
PassInGrid = NULL,
GridTune = FALSE,
MaxModelsInGrid = 30L,
MaxRunsWithoutNewWinner = 20L,
MaxRunMinutes = 60*60,
BaselineComparison = 'default',
# ML args
langevin = FALSE,
diffusion_temperature = 10000,
Trees = 1000,
Depth = 9,
L2_Leaf_Reg = NULL,
RandomStrength = 1,
BorderCount = 128,
LearningRate = NULL,
RSM = 1,
BootStrapType = NULL,
GrowPolicy = 'SymmetricTree',
model_size_reg = 0.5,
feature_border_type = 'GreedyLogSum',
sampling_unit = 'Object',
subsample = NULL,
score_function = 'Cosine',
min_data_in_leaf = 1)
# Insights Report
AutoQuant::ModelInsightsReport(
# Meta info
TargetColumnName = 'Adrian',
PredictionColumnName = 'Predict',
FeatureColumnNames = Features,
DateColumnName = NULL,
# Control options
TargetType = 'regression',
ModelID = 'Test_Model_1',
Algo = 'catboost',
OutputPath = getwd(),
ModelObject = TestModel)
# Score data
Preds <- AutoQuant::AutoCatBoostScoring(
TargetType = 'regression',
ScoringData = data,
FeatureColumnNames = Features,
FactorLevelsList = TestModel$FactorLevelsList,
IDcols = c('IDcol_1','IDcol_2'),
OneHot = FALSE,
ReturnShapValues = TRUE,
ModelObject = TestModel$Model,
ModelPath = NULL,
ModelID = 'Test_Model_1',
ReturnFeatures = TRUE,
MultiClassTargetLevels = NULL,
TransformNumeric = FALSE,
BackTransNumeric = FALSE,
TargetColumnName = NULL,
TransformationObject = NULL,
TransID = NULL,
TransPath = NULL,
MDP_Impute = TRUE,
MDP_CharToFactor = TRUE,
MDP_RemoveDates = TRUE,
MDP_MissFactor = '0',
MDP_MissNum = -1,
RemoveModel = FALSE)AutoCatBoostClassifier() Scoring Example
# Refresh data
data <- AutoQuant::FakeDataGenerator(
Correlation = 0.85,
N = 25000L,
ID = 2L,
AddWeightsColumn = TRUE,
ZIP = 0L,
AddDate = TRUE,
Classification = TRUE,
MultiClass = FALSE)
# Copy data (used for scoring below``)
data1 <- data.table::copy(data)
# Partition Data
Sets <- Rodeo::AutoDataPartition(
data = data,
NumDataSets = 3,
Ratios = c(0.7,0.2,0.1),
PartitionType = "random",
StratifyColumnNames = "Adrian",
TimeColumnName = NULL)
TTrainData <- Sets$TrainData
VValidationData <- Sets$ValidationData
TTestData <- Sets$TestData
rm(Sets)
# Feature Colnames
Features <- names(TTrainData)[!names(TTrainData) %in% c("IDcol_1", "IDcol_2","DateTime","Adrian")]
# AutoCatBoostClassifier
TestModel <- AutoQuant::AutoCatBoostClassifier(
# GPU or CPU and the number of available GPUs
task_type = "CPU",
NumGPUs = 1,
# Metadata arguments
OutputSelection = c("Importances", "EvalPlots", "EvalMetrics", "Score_TrainData"),
ModelID = "Test_Model_1",
model_path = normalizePath("./"),
metadata_path = normalizePath("./"),
SaveModelObjects = FALSE,
ReturnModelObjects = TRUE,
SaveInfoToPDF = FALSE,
# Data arguments
data = TTrainData,
TrainOnFull = FALSE,
ValidationData = VValidationData,
TestData = TTestData,
TargetColumnName = "Adrian",
FeatureColNames = Features,
PrimaryDateColumn = "DateTime",
WeightsColumnName = "Weights",
ClassWeights = c(1L,1L),
IDcols = c("IDcol_1","IDcol_2","DateTime"),
# Model evaluation
CostMatrixWeights = c(2,0,0,1),
EvalMetric = "MCC",
LossFunction = "Logloss",
grid_eval_metric = "Utility",
MetricPeriods = 10L,
NumOfParDepPlots = 3,
# Grid tuning arguments
PassInGrid = NULL,
GridTune = FALSE,
MaxModelsInGrid = 30L,
MaxRunsWithoutNewWinner = 20L,
MaxRunMinutes = 24L*60L,
BaselineComparison = "default",
# ML args
Trees = 100L,
Depth = 4L,
LearningRate = NULL,
L2_Leaf_Reg = NULL,
RandomStrength = 1,
BorderCount = 128,
RSM = 0.80,
BootStrapType = "Bayesian",
GrowPolicy = "SymmetricTree",
langevin = FALSE,
diffusion_temperature = 10000,
model_size_reg = 0.5,
feature_border_type = "GreedyLogSum",
sampling_unit = "Object",
subsample = NULL,
score_function = "Cosine",
min_data_in_leaf = 1,
DebugMode = TRUE)
# Insights Report
AutoQuant::ModelInsightsReport(
# Meta info
TargetColumnName = 'Adrian',
PredictionColumnName = 'p1',
FeatureColumnNames = Features,
DateColumnName = NULL,
# Control options
TargetType = 'classification',
ModelID = 'Test_Model_1',
Algo = 'catboost',
OutputPath = getwd(),
ModelObject = TestModel)
# Score data
Preds <- AutoQuant::AutoCatBoostScoring(
TargetType = 'classifier',
ScoringData = data,
FeatureColumnNames = Features,
FactorLevelsList = TestModel$FactorLevelsList,
IDcols = c("IDcol_1","IDcol_2","DateTime"),
OneHot = FALSE,
ReturnShapValues = TRUE,
ModelObject = TestModel$Model,
ModelPath = NULL,
ModelID = 'Test_Model_1',
ReturnFeatures = TRUE,
MultiClassTargetLevels = NULL,
TransformNumeric = FALSE,
BackTransNumeric = FALSE,
TargetColumnName = NULL,
TransformationObject = NULL,
TransID = NULL,
TransPath = NULL,
MDP_Impute = TRUE,
MDP_CharToFactor = TRUE,
MDP_RemoveDates = TRUE,
MDP_MissFactor = '0',
MDP_MissNum = -1,
RemoveModel = FALSE)AutoCatBoostMultiClasss() Scoring Example
# Refresh data
data <- AutoQuant::FakeDataGenerator(
Correlation = 0.85,
N = 25000L,
ID = 2L,
AddWeightsColumn = TRUE,
ZIP = 0L,
AddDate = TRUE,
Classification = FALSE,
MultiClass = TRUE)
# Copy data (used for scoring below``)
data1 <- data.table::copy(data)
# Partition Data
Sets <- Rodeo::AutoDataPartition(
data = data,
NumDataSets = 3,
Ratios = c(0.7,0.2,0.1),
PartitionType = "random",
StratifyColumnNames = "Adrian",
TimeColumnName = NULL)
TTrainData <- Sets$TrainData
VValidationData <- Sets$ValidationData
TTestData <- Sets$TestData
rm(Sets)
# Feature Colnames
Features <- names(TTrainData)[!names(TTrainData) %in% c("IDcol_1", "IDcol_2","Adrian","DateTime")]
# Run function
TestModel <- AutoQuant::AutoCatBoostMultiClass(
# GPU or CPU and the number of available GPUs
task_type = "GPU",
NumGPUs = 1,
# Metadata arguments
OutputSelection = c("Importances", "EvalPlots", "EvalMetrics", "Score_TrainData"),
ModelID = "Test_Model_1",
model_path = normalizePath("./"),
metadata_path = normalizePath("./"),
SaveModelObjects = FALSE,
ReturnModelObjects = TRUE,
# Data arguments
data = TTrainData,
TrainOnFull = FALSE,
ValidationData = VValidationData,
TestData = TTestData,
TargetColumnName = "Adrian",
FeatureColNames = Features,
PrimaryDateColumn = "DateTime",
WeightsColumnName = "Weights",
ClassWeights = c(1L,1L,1L,1L,1L),
IDcols = c("IDcol_1","IDcol_2","DateTime"),
# Model evaluation
eval_metric = "MCC",
loss_function = "MultiClassOneVsAll",
grid_eval_metric = "Accuracy",
MetricPeriods = 10L,
# Grid tuning arguments
PassInGrid = NULL,
GridTune = FALSE,
MaxModelsInGrid = 30L,
MaxRunsWithoutNewWinner = 20L,
MaxRunMinutes = 24L*60L,
BaselineComparison = "default",
# ML args
Trees = 100L,
Depth = 4L,
LearningRate = 0.01,
L2_Leaf_Reg = 1.0,
RandomStrength = 1,
BorderCount = 128,
langevin = FALSE,
diffusion_temperature = 10000,
RSM = 0.80,
BootStrapType = "Bayesian",
GrowPolicy = "SymmetricTree",
model_size_reg = 0.5,
feature_border_type = "GreedyLogSum",
sampling_unit = "Group",
subsample = NULL,
score_function = "Cosine",
min_data_in_leaf = 1,
DebugMode = TRUE)
# Insights Report
AutoQuant::ModelInsightsReport(
# Meta info
TargetColumnName = 'Adrian',
PredictionColumnName = 'Predict',
FeatureColumnNames = Features,
DateColumnName = NULL,
# Control options
TargetType = 'multiclass',
ModelID = 'Test_Model_1',
Algo = 'catboost',
OutputPath = getwd(),
ModelObject = TestModel)
# Score data
Preds <- AutoQuant::AutoCatBoostScoring(
TargetType = 'multiclass',
ScoringData = data,
FeatureColumnNames = Features,
FactorLevelsList = TestModel$FactorLevelsList,
IDcols = c("IDcol_1","IDcol_2","DateTime"),
OneHot = FALSE,
ReturnShapValues = FALSE,
ModelObject = TestModel$Model,
ModelPath = NULL,
ModelID = 'Test_Model_1',
ReturnFeatures = TRUE,
MultiClassTargetLevels = TestModel$TargetLevels,
TransformNumeric = FALSE,
BackTransNumeric = FALSE,
TargetColumnName = NULL,
TransformationObject = NULL,
TransID = NULL,
TransPath = NULL,
MDP_Impute = TRUE,
MDP_CharToFactor = TRUE,
MDP_RemoveDates = TRUE,
MDP_MissFactor = '0',
MDP_MissNum = -1,
RemoveModel = FALSE)AutoLightGBM__() Examples
AutoLightGBMRegression() Scoring Example
# Refresh data
data <- AutoQuant::FakeDataGenerator(
Correlation = 0.85,
N = 25000L,
ID = 2L,
AddWeightsColumn = TRUE,
ZIP = 0L,
AddDate = TRUE,
Classification = FALSE,
MultiClass = FALSE)
# Partition Data
Sets <- Rodeo::AutoDataPartition(
data = data,
NumDataSets = 3,
Ratios = c(0.7,0.2,0.1),
PartitionType = "random",
StratifyColumnNames = "Adrian",
TimeColumnName = NULL)
TTrainData <- Sets$TrainData
VValidationData <- Sets$ValidationData
TTestData <- Sets$TestData
rm(Sets)
# Run function
TestModel <- AutoQuant::AutoLightGBMRegression(
# GPU or CPU
NThreads = parallel::detectCores(),
# Metadata args
OutputSelection = c("Importances","EvalPlots","EvalMetrics","Score_TrainData"),
model_path = getwd(),
metadata_path = getwd(),
ModelID = "Test_Model_1",
NumOfParDepPlots = 3L,
EncodingMethod = "credibility",
ReturnFactorLevels = TRUE,
ReturnModelObjects = TRUE,
SaveModelObjects = TRUE,
SaveInfoToPDF = FALSE,
DebugMode = TRUE,
# Data args
data = TTrainData,
TrainOnFull = FALSE,
ValidationData = VValidationData,
TestData = TTestData,
TargetColumnName = "Adrian",
FeatureColNames = names(TTrainData)[!names(TTrainData) %in% c("IDcol_1", "IDcol_2","DateTime","Adrian")],
PrimaryDateColumn = "DateTime",
WeightsColumnName = "Weights",
IDcols = c("IDcol_1","IDcol_2","DateTime"),
TransformNumericColumns = NULL,
Methods = c("Asinh","Asin","Log","LogPlus1","Sqrt","Logit"),
# Grid parameters
GridTune = FALSE,
grid_eval_metric = "r2",
BaselineComparison = "default",
MaxModelsInGrid = 10L,
MaxRunsWithoutNewWinner = 20L,
MaxRunMinutes = 24L*60L,
PassInGrid = NULL,
# Core parameters
# https://lightgbm.readthedocs.io/en/latest/Parameters.html#core-parameters
input_model = NULL, # continue training a model that is stored to file
task = "train",
device_type = "CPU",
objective = 'regression',
metric = "rmse",
boosting = "gbdt",
LinearTree = FALSE,
Trees = 50L,
eta = NULL,
num_leaves = 31,
deterministic = TRUE,
# Learning Parameters
# https://lightgbm.readthedocs.io/en/latest/Parameters.html#learning-control-parameters
force_col_wise = FALSE,
force_row_wise = FALSE,
max_depth = 6,
min_data_in_leaf = 20,
min_sum_hessian_in_leaf = 0.001,
bagging_freq = 1.0,
bagging_fraction = 1.0,
feature_fraction = 1.0,
feature_fraction_bynode = 1.0,
lambda_l1 = 0.0,
lambda_l2 = 0.0,
extra_trees = FALSE,
early_stopping_round = 10,
first_metric_only = TRUE,
max_delta_step = 0.0,
linear_lambda = 0.0,
min_gain_to_split = 0,
drop_rate_dart = 0.10,
max_drop_dart = 50,
skip_drop_dart = 0.50,
uniform_drop_dart = FALSE,
top_rate_goss = FALSE,
other_rate_goss = FALSE,
monotone_constraints = NULL,
monotone_constraints_method = "advanced",
monotone_penalty = 0.0,
forcedsplits_filename = NULL, # use for AutoStack option; .json file
refit_decay_rate = 0.90,
path_smooth = 0.0,
# IO Dataset Parameters
# https://lightgbm.readthedocs.io/en/latest/Parameters.html#io-parameters
max_bin = 255,
min_data_in_bin = 3,
data_random_seed = 1,
is_enable_sparse = TRUE,
enable_bundle = TRUE,
use_missing = TRUE,
zero_as_missing = FALSE,
two_round = FALSE,
# Convert Parameters
convert_model = NULL,
convert_model_language = "cpp",
# Objective Parameters
# https://lightgbm.readthedocs.io/en/latest/Parameters.html#objective-parameters
boost_from_average = TRUE,
alpha = 0.90,
fair_c = 1.0,
poisson_max_delta_step = 0.70,
tweedie_variance_power = 1.5,
lambdarank_truncation_level = 30,
# Metric Parameters (metric is in Core)
# https://lightgbm.readthedocs.io/en/latest/Parameters.html#metric-parameters
is_provide_training_metric = TRUE,
eval_at = c(1,2,3,4,5),
# Network Parameters
# https://lightgbm.readthedocs.io/en/latest/Parameters.html#network-parameters
num_machines = 1,
# GPU Parameters
# https://lightgbm.readthedocs.io/en/latest/Parameters.html#gpu-parameters
gpu_platform_id = -1,
gpu_device_id = -1,
gpu_use_dp = TRUE,
num_gpu = 1)
# Outcome
ModelID = "Test_Model_1"
colnames <- data.table::fread(file = file.path(getwd(), paste0(ModelID, "_ColNames.csv")))
Preds <- AutoQuant::AutoLightGBMScoring(
TargetType = "regression",
ScoringData = TTestData,
ReturnShapValues = FALSE,
FeatureColumnNames = colnames[[1L]],
IDcols = c("IDcol_1","IDcol_2"),
EncodingMethod = "credibility",
FactorLevelsList = NULL,
TargetLevels = NULL,
ModelObject = NULL,
ModelPath = getwd(),
ModelID = "Test_Model_1",
ReturnFeatures = TRUE,
TransformNumeric = FALSE,
BackTransNumeric = FALSE,
TargetColumnName = NULL,
TransformationObject = NULL,
TransID = NULL,
TransPath = NULL,
MDP_Impute = TRUE,
MDP_CharToFactor = TRUE,
MDP_RemoveDates = TRUE,
MDP_MissFactor = "0",
MDP_MissNum = -1)AutoLightGBMClassifier() Scoring Example
# Refresh data
data <- AutoQuant::FakeDataGenerator(
Correlation = 0.85,
N = 25000L,
ID = 2L,
AddWeightsColumn = TRUE,
ZIP = 0L,
AddDate = TRUE,
Classification = TRUE,
MultiClass = FALSE)
# Partition Data
Sets <- Rodeo::AutoDataPartition(
data = data,
NumDataSets = 3,
Ratios = c(0.7,0.2,0.1),
PartitionType = "random",
StratifyColumnNames = "Adrian",
TimeColumnName = NULL)
TTrainData <- Sets$TrainData
VValidationData <- Sets$ValidationData
TTestData <- Sets$TestData
rm(Sets)
# Run function
TestModel <- AutoQuant::AutoLightGBMClassifier(
# Multithreading
NThreads = parallel::detectCores(),
# Metadata args
OutputSelection = c("Importances","EvalPlots","EvalMetrics","Score_TrainData"),
model_path = normalizePath("./"),
metadata_path = NULL,
ModelID = "Test_Model_1",
NumOfParDepPlots = 3L,
EncodingMethod = "credibility",
ReturnFactorLevels = TRUE,
ReturnModelObjects = TRUE,
SaveModelObjects = TRUE,
SaveInfoToPDF = FALSE,
DebugMode = TRUE,
# Data args
data = TTrainData,
TrainOnFull = FALSE,
ValidationData = VValidationData,
TestData = TTestData,
TargetColumnName = "Adrian",
FeatureColNames = names(TTrainData)[!names(TTrainData) %in% c("IDcol_1", "IDcol_2","DateTime","Adrian")],
PrimaryDateColumn = NULL,
WeightsColumnName = "Weights",
IDcols = c("IDcol_1","IDcol_2","DateTime"),
CostMatrixWeights = c(1,0,0,1),
# Grid parameters
GridTune = FALSE,
grid_eval_metric = "Utility",
BaselineComparison = "default",
MaxModelsInGrid = 10L,
MaxRunsWithoutNewWinner = 20L,
MaxRunMinutes = 24L*60L,
PassInGrid = NULL,
# Core parameters
# https://lightgbm.readthedocs.io/en/latest/Parameters.html#core-parameters
input_model = NULL, # continue training a model that is stored to file
task = "train",
device_type = "CPU",
objective = 'binary',
metric = 'binary_logloss',
boosting = "gbdt",
LinearTree = FALSE,
Trees = 50L,
eta = NULL,
num_leaves = 31,
deterministic = TRUE,
# Learning Parameters
# https://lightgbm.readthedocs.io/en/latest/Parameters.html#learning-control-parameters
force_col_wise = FALSE,
force_row_wise = FALSE,
max_depth = 6,
min_data_in_leaf = 20,
min_sum_hessian_in_leaf = 0.001,
bagging_freq = 1.0,
bagging_fraction = 1.0,
feature_fraction = 1.0,
feature_fraction_bynode = 1.0,
lambda_l1 = 0.0,
lambda_l2 = 0.0,
extra_trees = FALSE,
early_stopping_round = 10,
first_metric_only = TRUE,
max_delta_step = 0.0,
linear_lambda = 0.0,
min_gain_to_split = 0,
drop_rate_dart = 0.10,
max_drop_dart = 50,
skip_drop_dart = 0.50,
uniform_drop_dart = FALSE,
top_rate_goss = FALSE,
other_rate_goss = FALSE,
monotone_constraints = NULL,
monotone_constraints_method = 'advanced',
monotone_penalty = 0.0,
forcedsplits_filename = NULL, # use for AutoStack option; .json file
refit_decay_rate = 0.90,
path_smooth = 0.0,
# IO Dataset Parameters
# https://lightgbm.readthedocs.io/en/latest/Parameters.html#io-parameters
max_bin = 255,
min_data_in_bin = 3,
data_random_seed = 1,
is_enable_sparse = TRUE,
enable_bundle = TRUE,
use_missing = TRUE,
zero_as_missing = FALSE,
two_round = FALSE,
# Convert Parameters
convert_model = NULL,
convert_model_language = "cpp",
# Objective Parameters
# https://lightgbm.readthedocs.io/en/latest/Parameters.html#objective-parameters
boost_from_average = TRUE,
is_unbalance = FALSE,
scale_pos_weight = 1.0,
# Metric Parameters (metric is in Core)
# https://lightgbm.readthedocs.io/en/latest/Parameters.html#metric-parameters
is_provide_training_metric = TRUE,
eval_at = c(1,2,3,4,5),
# Network Parameters
# https://lightgbm.readthedocs.io/en/latest/Parameters.html#network-parameters
num_machines = 1,
# GPU Parameters
# https://lightgbm.readthedocs.io/en/latest/Parameters.html#gpu-parameters
gpu_platform_id = -1,
gpu_device_id = -1,
gpu_use_dp = TRUE,
num_gpu = 1)
# Outcome
ModelID = "Test_Model_1"
colnames <- data.table::fread(file = file.path(getwd(), paste0(ModelID, "_ColNames.csv")))
Preds <- AutoQuant::AutoLightGBMScoring(
TargetType = "classification",
ScoringData = TTestData,
ReturnShapValues = FALSE,
FeatureColumnNames = colnames[[1L]],
IDcols = c("IDcol_1","IDcol_2","DateTime"),
EncodingMethod = "credibility",
FactorLevelsList = NULL,
TargetLevels = NULL,
ModelObject = NULL,
ModelPath = getwd(),
ModelID = "Test_Model_1",
ReturnFeatures = TRUE,
TransformNumeric = FALSE,
BackTransNumeric = FALSE,
TargetColumnName = NULL,
TransformationObject = NULL,
TransID = NULL,
TransPath = NULL,
MDP_Impute = TRUE,
MDP_CharToFactor = TRUE,
MDP_RemoveDates = TRUE,
MDP_MissFactor = "0",
MDP_MissNum = -1)AutoLightGBMMultiClasss() Scoring Example
# Refresh data
data <- AutoQuant::FakeDataGenerator(
Correlation = 0.85,
N = 25000L,
ID = 2L,
AddWeightsColumn = TRUE,
ZIP = 0L,
AddDate = TRUE,
Classification = FALSE,
MultiClass = TRUE)
# Partition Data
Sets <- Rodeo::AutoDataPartition(
data = data,
NumDataSets = 3,
Ratios = c(0.7,0.2,0.1),
PartitionType = "random",
StratifyColumnNames = "Adrian",
TimeColumnName = NULL)
TTrainData <- Sets$TrainData
VValidationData <- Sets$ValidationData
TTestData <- Sets$TestData
rm(Sets)
# Run function
TestModel <- AutoQuant::AutoLightGBMMultiClass(
# GPU or CPU
NThreads = parallel::detectCores(),
# Metadata args
OutputSelection = c("Importances","EvalPlots","EvalMetrics","Score_TrainData"),
model_path = normalizePath("./"),
metadata_path = NULL,
ModelID = "Test_Model_1",
NumOfParDepPlots = 3L,
EncodingMethod = "credibility",
ReturnFactorLevels = TRUE,
ReturnModelObjects = TRUE,
SaveModelObjects = TRUE,
SaveInfoToPDF = FALSE,
DebugMode = TRUE,
# Data args
data = TTrainData,
TrainOnFull = FALSE,
ValidationData = VValidationData,
TestData = TTestData,
TargetColumnName = "Adrian",
FeatureColNames = names(TTrainData)[!names(TTrainData) %in% c("IDcol_1","IDcol_2","DateTime","Adrian")],
PrimaryDateColumn = NULL,
WeightsColumnName = "Weights",
IDcols = c("IDcol_1","IDcol_2",'DateTime'),
CostMatrixWeights = c(1,0,0,1),
# Grid parameters
GridTune = FALSE,
grid_eval_metric = "microauc",
BaselineComparison = "default",
MaxModelsInGrid = 10L,
MaxRunsWithoutNewWinner = 20L,
MaxRunMinutes = 24L*60L,
PassInGrid = NULL,
# Core parameters
# https://lightgbm.readthedocs.io/en/latest/Parameters.html#core-parameters
input_model = NULL, # continue training a model that is stored to file
task = "train",
device_type = "CPU",
objective = 'multiclass',
multi_error_top_k = 1,
metric = 'multiclass_logloss',
boosting = "gbdt",
LinearTree = FALSE,
Trees = 50L,
eta = NULL,
num_leaves = 31,
deterministic = TRUE,
# Learning Parameters
# https://lightgbm.readthedocs.io/en/latest/Parameters.html#learning-control-parameters
force_col_wise = FALSE,
force_row_wise = FALSE,
max_depth = 6,
min_data_in_leaf = 20,
min_sum_hessian_in_leaf = 0.001,
bagging_freq = 1.0,
bagging_fraction = 1.0,
feature_fraction = 1.0,
feature_fraction_bynode = 1.0,
lambda_l1 = 0.0,
lambda_l2 = 0.0,
extra_trees = FALSE,
early_stopping_round = 10,
first_metric_only = TRUE,
max_delta_step = 0.0,
linear_lambda = 0.0,
min_gain_to_split = 0,
drop_rate_dart = 0.10,
max_drop_dart = 50,
skip_drop_dart = 0.50,
uniform_drop_dart = FALSE,
top_rate_goss = FALSE,
other_rate_goss = FALSE,
monotone_constraints = NULL,
monotone_constraints_method = 'advanced',
monotone_penalty = 0.0,
forcedsplits_filename = NULL, # use for AutoStack option; .json file
refit_decay_rate = 0.90,
path_smooth = 0.0,
# IO Dataset Parameters
# https://lightgbm.readthedocs.io/en/latest/Parameters.html#io-parameters
max_bin = 255,
min_data_in_bin = 3,
data_random_seed = 1,
is_enable_sparse = TRUE,
enable_bundle = TRUE,
use_missing = TRUE,
zero_as_missing = FALSE,
two_round = FALSE,
# Convert Parameters
convert_model = NULL,
convert_model_language = "cpp",
# Objective Parameters
# https://lightgbm.readthedocs.io/en/latest/Parameters.html#objective-parameters
boost_from_average = TRUE,
is_unbalance = FALSE,
scale_pos_weight = 1.0,
# Metric Parameters (metric is in Core)
# https://lightgbm.readthedocs.io/en/latest/Parameters.html#metric-parameters
is_provide_training_metric = TRUE,
eval_at = c(1,2,3,4,5),
# Network Parameters
# https://lightgbm.readthedocs.io/en/latest/Parameters.html#network-parameters
num_machines = 1,
# GPU Parameters
# https://lightgbm.readthedocs.io/en/latest/Parameters.html#gpu-parameters
gpu_platform_id = -1,
gpu_device_id = -1,
gpu_use_dp = TRUE,
num_gpu = 1)
# Outcome
ModelID = "Test_Model_1"
colnames <- data.table::fread(file = file.path(getwd(), paste0(ModelID, "_ColNames.csv")))
Preds <- AutoQuant::AutoLightGBMScoring(
TargetType = "multiclass",
ScoringData = TTestData,
ReturnShapValues = FALSE,
FeatureColumnNames = colnames[[1L]],
IDcols = c("IDcol_1","IDcol_2","DateTime"),
EncodingMethod = "credibility",
FactorLevelsList = NULL,
TargetLevels = NULL,
ModelObject = NULL,
ModelPath = getwd(),
ModelID = "Test_Model_1",
ReturnFeatures = TRUE,
TransformNumeric = FALSE,
BackTransNumeric = FALSE,
TargetColumnName = NULL,
TransformationObject = NULL,
TransID = NULL,
TransPath = NULL,
MDP_Impute = TRUE,
MDP_CharToFactor = TRUE,
MDP_RemoveDates = TRUE,
MDP_MissFactor = "0",
MDP_MissNum = -1)AutoLightGBMHurdleModel() Scoring Example
# Classify
Classify <- TRUE
# Get data
if(Classify) {
data <- AutoQuant::FakeDataGenerator(N = 15000, ZIP = 1)
} else {
data <- AutoQuant::FakeDataGenerator(N = 100000, ZIP = 2)
}
# Partition Data
Sets <- Rodeo::AutoDataPartition(
data = data,
NumDataSets = 3,
Ratios = c(0.7,0.2,0.1),
PartitionType = "random",
StratifyColumnNames = "Adrian",
TimeColumnName = NULL)
TTrainData <- Sets$TrainData
VValidationData <- Sets$ValidationData
TTestData <- Sets$TestData
rm(Sets)
# Run function
TestModel <- AutoQuant::AutoLightGBMHurdleModel(
# Operationalization
ModelID = 'ModelTest',
SaveModelObjects = FALSE,
ReturnModelObjects = TRUE,
NThreads = parallel::detectCores(),
# Data related args
data = TTrainData,
ValidationData = VValidationData,
PrimaryDateColumn = "DateTime",
TestData = TTestData,
WeightsColumnName = NULL,
TrainOnFull = FALSE,
Buckets = if(Classify) 0L else c(0,2,3),
TargetColumnName = "Adrian",
FeatureColNames = names(TTrainData)[!names(data) %in% c("Adrian","IDcol_1","IDcol_2","IDcol_3","IDcol_4","IDcol_5","DateTime")],
IDcols = c("IDcol_1","IDcol_2","IDcol_3","IDcol_4","IDcol_5","DateTime"),
DebugMode = TRUE,
# Metadata args
EncodingMethod = "credibility",
Paths = getwd(),
MetaDataPaths = NULL,
TransformNumericColumns = NULL,
Methods = c('Asinh', 'Asin', 'Log', 'LogPlus1', 'Logit'),
ClassWeights = c(1,1),
SplitRatios = NULL,
NumOfParDepPlots = 10L,
# Grid tuning setup
PassInGrid = NULL,
GridTune = FALSE,
BaselineComparison = 'default',
MaxModelsInGrid = 1L,
MaxRunsWithoutNewWinner = 20L,
MaxRunMinutes = 60L*60L,
# LightGBM parameters
task = list('classifier' = 'train', 'regression' = 'train'),
device_type = list('classifier' = 'CPU', 'regression' = 'CPU'),
objective = if(Classify) list('classifier' = 'binary', 'regression' = 'regression') else list('classifier' = 'multiclass', 'regression' = 'regression'),
metric = if(Classify) list('classifier' = 'binary_logloss', 'regression' = 'rmse') else list('classifier' = 'multi_logloss', 'regression' = 'rmse'),
boosting = list('classifier' = 'gbdt', 'regression' = 'gbdt'),
LinearTree = list('classifier' = FALSE, 'regression' = FALSE),
Trees = list('classifier' = 50L, 'regression' = 50L),
eta = list('classifier' = NULL, 'regression' = NULL),
num_leaves = list('classifier' = 31, 'regression' = 31),
deterministic = list('classifier' = TRUE, 'regression' = TRUE),
# Learning Parameters
force_col_wise = list('classifier' = FALSE, 'regression' = FALSE),
force_row_wise = list('classifier' = FALSE, 'regression' = FALSE),
max_depth = list('classifier' = NULL, 'regression' = NULL),
min_data_in_leaf = list('classifier' = 20, 'regression' = 20),
min_sum_hessian_in_leaf = list('classifier' = 0.001, 'regression' = 0.001),
bagging_freq = list('classifier' = 0, 'regression' = 0),
bagging_fraction = list('classifier' = 1.0, 'regression' = 1.0),
feature_fraction = list('classifier' = 1.0, 'regression' = 1.0),
feature_fraction_bynode = list('classifier' = 1.0, 'regression' = 1.0),
extra_trees = list('classifier' = FALSE, 'regression' = FALSE),
early_stopping_round = list('classifier' = 10, 'regression' = 10),
first_metric_only = list('classifier' = TRUE, 'regression' = TRUE),
max_delta_step = list('classifier' = 0.0, 'regression' = 0.0),
lambda_l1 = list('classifier' = 0.0, 'regression' = 0.0),
lambda_l2 = list('classifier' = 0.0, 'regression' = 0.0),
linear_lambda = list('classifier' = 0.0, 'regression' = 0.0),
min_gain_to_split = list('classifier' = 0, 'regression' = 0),
drop_rate_dart = list('classifier' = 0.10, 'regression' = 0.10),
max_drop_dart = list('classifier' = 50, 'regression' = 50),
skip_drop_dart = list('classifier' = 0.50, 'regression' = 0.50),
uniform_drop_dart = list('classifier' = FALSE, 'regression' = FALSE),
top_rate_goss = list('classifier' = FALSE, 'regression' = FALSE),
other_rate_goss = list('classifier' = FALSE, 'regression' = FALSE),
monotone_constraints = list('classifier' = NULL, 'regression' = NULL),
monotone_constraints_method = list('classifier' = 'advanced', 'regression' = 'advanced'),
monotone_penalty = list('classifier' = 0.0, 'regression' = 0.0),
forcedsplits_filename = list('classifier' = NULL, 'regression' = NULL),
refit_decay_rate = list('classifier' = 0.90, 'regression' = 0.90),
path_smooth = list('classifier' = 0.0, 'regression' = 0.0),
# IO Dataset Parameters
max_bin = list('classifier' = 255, 'regression' = 255),
min_data_in_bin = list('classifier' = 3, 'regression' = 3),
data_random_seed = list('classifier' = 1, 'regression' = 1),
is_enable_sparse = list('classifier' = TRUE, 'regression' = TRUE),
enable_bundle = list('classifier' = TRUE, 'regression' = TRUE),
use_missing = list('classifier' = TRUE, 'regression' = TRUE),
zero_as_missing = list('classifier' = FALSE, 'regression' = FALSE),
two_round = list('classifier' = FALSE, 'regression' = FALSE),
# Convert Parameters
convert_model = list('classifier' = NULL, 'regression' = NULL),
convert_model_language = list('classifier' = "cpp", 'regression' = "cpp"),
# Objective Parameters
boost_from_average = list('classifier' = TRUE, 'regression' = TRUE),
is_unbalance = list('classifier' = FALSE, 'regression' = FALSE),
scale_pos_weight = list('classifier' = 1.0, 'regression' = 1.0),
# Metric Parameters (metric is in Core)
is_provide_training_metric = list('classifier' = TRUE, 'regression' = TRUE),
eval_at = list('classifier' = c(1,2,3,4,5), 'regression' = c(1,2,3,4,5)),
# Network Parameters
num_machines = list('classifier' = 1, 'regression' = 1),
# GPU Parameters
gpu_platform_id = list('classifier' = -1, 'regression' = -1),
gpu_device_id = list('classifier' = -1, 'regression' = -1),
gpu_use_dp = list('classifier' = TRUE, 'regression' = TRUE),
num_gpu = list('classifier' = 1, 'regression' = 1))
# Remove Target Variable
TTrainData[, c("Target_Buckets", "Adrian") := NULL]
# Score LightGBM Hurdle Model
Output <- AutoQuant::AutoLightGBMHurdleModelScoring(
TestData = TTrainData,
Path = NULL,
ModelID = "ModelTest",
ModelList = TestModel$ModelList,
ArgsList = TestModel$ArgsList,
Threshold = NULL)AutoXGBoost__() Examples
AutoXGBoostRegression() Scoring Example
# Refresh data
data <- AutoQuant::FakeDataGenerator(
Correlation = 0.85,
N = 25000L,
ID = 2L,
FactorCount = 3,
AddWeightsColumn = TRUE,
ZIP = 0L,
AddDate = TRUE,
Classification = FALSE,
MultiClass = FALSE)
# Copy data
data1 <- data.table::copy(data)
# Partition Data
Sets <- Rodeo::AutoDataPartition(
data = data1,
NumDataSets = 3,
Ratios = c(0.7,0.2,0.1),
PartitionType = "random",
StratifyColumnNames = "Adrian",
TimeColumnName = NULL)
TTrainData <- Sets$TrainData
VValidationData <- Sets$ValidationData
TTestData <- Sets$TestData
rm(Sets)
# Run function
TestModel <- AutoQuant::AutoXGBoostRegression(
# GPU or CPU
TreeMethod = "hist",
NThreads = parallel::detectCores(),
LossFunction = 'reg:squarederror',
# Metadata arguments
model_path = normalizePath("./"),
metadata_path = NULL,
ModelID = "Test_Model_1",
EncodingMethod = "credibility",
ReturnFactorLevels = TRUE,
ReturnModelObjects = TRUE,
SaveModelObjects = TRUE,
DebugMode = TRUE,
# Data arguments
data = TTrainData,
TrainOnFull = FALSE,
ValidationData = VValidationData,
TestData = TTestData,
TargetColumnName = "Adrian",
FeatureColNames = names(TTrainData)[!names(TTrainData) %in% c("IDcol_1", "IDcol_2","DateTime","Adrian")],
WeightsColumnName = "Weights",
IDcols = c("IDcol_1","IDcol_2","DateTime"),
TransformNumericColumns = NULL,
Methods = c("Asinh", "Asin", "Log", "LogPlus1", "Sqrt", "Logit"),
# Model evaluation
eval_metric = "rmse",
NumOfParDepPlots = 3L,
# Grid tuning arguments
PassInGrid = NULL,
GridTune = FALSE,
grid_eval_metric = "r2",
BaselineComparison = "default",
MaxModelsInGrid = 10L,
MaxRunsWithoutNewWinner = 20L,
MaxRunMinutes = 24L*60L,
Verbose = 1L,
SaveInfoToPDF = TRUE,
# ML args
Trees = 50L,
eta = 0.05,
max_depth = 4L,
min_child_weight = 1.0,
subsample = 0.55,
colsample_bytree = 0.55)
# Score model
Preds <- AutoQuant::AutoXGBoostScoring(
TargetType = "regression",
ScoringData = data,
ReturnShapValues = FALSE,
FeatureColumnNames = names(TTrainData)[!names(TTrainData) %in% c("IDcol_1", "IDcol_2","DateTime","Adrian")],
IDcols = c("IDcol_1","IDcol_2","DateTime"),
EncodingMethod = "credibility",
FactorLevelsList = TestModel$FactorLevelsList,
TargetLevels = NULL,
ModelObject = TestModel$Model,
ModelPath = "home",
ModelID = "ModelTest",
ReturnFeatures = TRUE,
TransformNumeric = FALSE,
BackTransNumeric = FALSE,
TargetColumnName = NULL,
TransformationObject = NULL,
TransID = NULL,
TransPath = NULL,
MDP_Impute = TRUE,
MDP_CharToFactor = TRUE,
MDP_RemoveDates = TRUE,
MDP_MissFactor = "0",
MDP_MissNum = -1)AutoXGBoostClassifier() Scoring Example
# Refresh data
data <- AutoQuant::FakeDataGenerator(
Correlation = 0.85,
N = 25000L,
ID = 2L,
AddWeightsColumn = TRUE,
ZIP = 0L,
AddDate = TRUE,
Classification = TRUE,
MultiClass = FALSE)
# Partition Data
Sets <- Rodeo::AutoDataPartition(
data = data,
NumDataSets = 3,
Ratios = c(0.7,0.2,0.1),
PartitionType = "random",
StratifyColumnNames = "Adrian",
TimeColumnName = NULL)
TTrainData <- Sets$TrainData
VValidationData <- Sets$ValidationData
TTestData <- Sets$TestData
rm(Sets)
# Run function
TestModel <- AutoQuant::AutoXGBoostClassifier(
# GPU or CPU
TreeMethod = "hist",
NThreads = parallel::detectCores(),
# Metadata arguments
OutputSelection = c("Importances", "EvalPlots", "EvalMetrics", "Score_TrainData"),
model_path = normalizePath("./"),
metadata_path = NULL,
ModelID = "Test_Model_1",
EncodingMethod = "credibility",
ReturnFactorLevels = TRUE,
ReturnModelObjects = TRUE,
SaveModelObjects = TRUE,
SaveInfoToPDF = TRUE,
DebugMode = TRUE,
# Data arguments
data = TTrainData,
TrainOnFull = FALSE,
ValidationData = VValidationData,
TestData = TTestData,
TargetColumnName = "Adrian",
FeatureColNames = names(TTrainData)[!names(TTrainData) %in% c("IDcol_1", "IDcol_2","DateTime","Adrian")],
WeightsColumnName = "Weights",
IDcols = c("IDcol_1","IDcol_2","DateTime"),
# Model evaluation
LossFunction = 'reg:logistic',
eval_metric = "auc",
grid_eval_metric = "MCC",
CostMatrixWeights = c(1,0,0,1),
NumOfParDepPlots = 3L,
# Grid tuning arguments
PassInGrid = NULL,
GridTune = FALSE,
BaselineComparison = "default",
MaxModelsInGrid = 10L,
MaxRunsWithoutNewWinner = 20L,
MaxRunMinutes = 24L*60L,
Verbose = 1L,
# ML Args
Trees = 50L,
eta = 0.05,
max_depth = 4L,
min_child_weight = 1.0,
subsample = 0.55,
colsample_bytree = 0.55)
# Score model
Preds <- AutoQuant::AutoXGBoostScoring(
TargetType = "classifier",
ScoringData = data,
ReturnShapValues = FALSE,
FeatureColumnNames = names(TTrainData)[!names(TTrainData) %in% c("IDcol_1", "IDcol_2","DateTime","Adrian")],
IDcols = c("IDcol_1","IDcol_2","DateTime"),
EncodingMethod = "credibility",
FactorLevelsList = TestModel$FactorLevelsList,
TargetLevels = NULL,
ModelObject = TestModel$Model,
ModelPath = "home",
ModelID = "ModelTest",
ReturnFeatures = TRUE,
TransformNumeric = FALSE,
BackTransNumeric = FALSE,
TargetColumnName = NULL,
TransformationObject = NULL,
TransID = NULL,
TransPath = NULL,
MDP_Impute = TRUE,
MDP_CharToFactor = TRUE,
MDP_RemoveDates = TRUE,
MDP_MissFactor = "0",
MDP_MissNum = -1)AutoXGBoostMultiClasss() Scoring Example
# Refresh data
data <- AutoQuant::FakeDataGenerator(
Correlation = 0.85,
N = 25000L,
ID = 2L,
AddWeightsColumn = TRUE,
ZIP = 0L,
AddDate = TRUE,
Classification = FALSE,
MultiClass = TRUE)
# Partition Data
Sets <- Rodeo::AutoDataPartition(
data = data,
NumDataSets = 3,
Ratios = c(0.7,0.2,0.1),
PartitionType = "random",
StratifyColumnNames = "Adrian",
TimeColumnName = NULL)
TTrainData <- Sets$TrainData
VValidationData <- Sets$ValidationData
TTestData <- Sets$TestData
rm(Sets)
# Run function
TestModel <- AutoQuant::AutoXGBoostMultiClass(
# GPU or CPU
TreeMethod = "hist",
NThreads = parallel::detectCores(),
# Metadata arguments
OutputSelection = c("Importances", "EvalPlots", "EvalMetrics", "Score_TrainData"),
model_path = normalizePath("./"),
metadata_path = normalizePath("./"),
ModelID = "Test_Model_1",
ReturnFactorLevels = TRUE,
ReturnModelObjects = TRUE,
SaveModelObjects = FALSE,
EncodingMethod = "credibility",
DebugMode = TRUE,
# Data arguments
data = TTrainData,
TrainOnFull = FALSE,
ValidationData = VValidationData,
TestData = TTestData,
TargetColumnName = "Adrian",
FeatureColNames = names(TTrainData)[!names(TTrainData) %in% c("IDcol_1", "IDcol_2","DateTime","Adrian")],
IDcols = c("IDcol_1","IDcol_2","DateTime"),
# Model evaluation
eval_metric = "merror",
LossFunction = 'multi:softprob',
grid_eval_metric = "accuracy",
NumOfParDepPlots = 3L,
# Grid tuning arguments
PassInGrid = NULL,
GridTune = FALSE,
BaselineComparison = "default",
MaxModelsInGrid = 10L,
MaxRunsWithoutNewWinner = 20L,
MaxRunMinutes = 24L*60L,
Verbose = 1L,
# ML Args
Trees = 50L,
eta = 0.05,
max_depth = 4L,
min_child_weight = 1.0,
subsample = 0.55,
colsample_bytree = 0.55)
# Score model
Preds <- AutoQuant::AutoXGBoostScoring(
TargetType = "multiclass",
ScoringData = data,
ReturnShapValues = FALSE,
FeatureColumnNames = names(TTrainData)[!names(TTrainData) %in% c("IDcol_1", "IDcol_2","DateTime","Adrian")],
IDcols = c("IDcol_1","IDcol_2","DateTime"),
EncodingMethod = "credibility",
FactorLevelsList = TestModel$FactorLevelsList,
TargetLevels = TestModel$TargetLevels,
ModelObject = TestModel$Model,
ModelPath = NULL,
ModelID = "ModelTest",
ReturnFeatures = TRUE,
TransformNumeric = FALSE,
BackTransNumeric = FALSE,
TargetColumnName = NULL,
TransformationObject = NULL,
TransID = NULL,
TransPath = NULL,
MDP_Impute = TRUE,
MDP_CharToFactor = TRUE,
MDP_RemoveDates = TRUE,
MDP_MissFactor = "0",
MDP_MissNum = -1)AutoXGBoostHurdleModel() Scoring Example
# Classify
Classify <- TRUE
# Get data
if(Classify) {
data <- AutoQuant::FakeDataGenerator(N = 15000, ZIP = 1)
} else {
data <- AutoQuant::FakeDataGenerator(N = 100000, ZIP = 2)
}
# Partition Data
Sets <- Rodeo::AutoDataPartition(
data = data,
NumDataSets = 3,
Ratios = c(0.7,0.2,0.1),
PartitionType = "random",
StratifyColumnNames = "Adrian",
TimeColumnName = NULL)
TTrainData <- Sets$TrainData
VValidationData <- Sets$ValidationData
TTestData <- Sets$TestData
rm(Sets)
# Run function
TestModel <- AutoQuant::AutoXGBoostHurdleModel(
# Operationalization
ModelID = 'ModelTest',
SaveModelObjects = FALSE,
ReturnModelObjects = TRUE,
NThreads = parallel::detectCores(),
# Data related args
data = TTrainData,
ValidationData = VValidationData,
PrimaryDateColumn = "DateTime",
TestData = TTestData,
WeightsColumnName = NULL,
TrainOnFull = FALSE,
Buckets = if(Classify) 0L else c(0,2,3),
TargetColumnName = "Adrian",
FeatureColNames = names(TTrainData)[!names(data) %in% c("Adrian","IDcol_1","IDcol_2","IDcol_3","IDcol_4","IDcol_5","DateTime")],
IDcols = c("IDcol_1","IDcol_2","IDcol_3","IDcol_4","IDcol_5","DateTime"),
DebugMode = FALSE,
# Metadata args
EncodingMethod = "credibility",
Paths = normalizePath('./'),
MetaDataPaths = NULL,
TransformNumericColumns = NULL,
Methods = c('Asinh', 'Asin', 'Log', 'LogPlus1', 'Logit'),
ClassWeights = c(1,1),
SplitRatios = NULL,
NumOfParDepPlots = 10L,
# Grid tuning setup
PassInGrid = NULL,
GridTune = FALSE,
BaselineComparison = 'default',
MaxModelsInGrid = 1L,
MaxRunsWithoutNewWinner = 20L,
MaxRunMinutes = 60L*60L,
# XGBoost parameters
TreeMethod = "hist",
Trees = list("classifier" = 50, "regression" = 50),
eta = list("classifier" = 0.05, "regression" = 0.05),
max_depth = list("classifier" = 4L, "regression" = 4L),
min_child_weight = list("classifier" = 1.0, "regression" = 1.0),
subsample = list("classifier" = 0.55, "regression" = 0.55),
colsample_bytree = list("classifier" = 0.55, "regression" = 0.55))
# Remove Target Variable
TTrainData[, c("Target_Buckets", "Adrian") := NULL]
# Score XGBoost Hurdle Model
Output <- AutoQuant::AutoXGBoostHurdleModelScoring(
TestData = TTrainData,
Path = NULL,
ModelID = "ModelTest",
ModelList = TestModel$ModelList,
ArgsList = TestModel$ArgsList,
Threshold = NULL)Expand to view content
Regression ModelInsightsReport() Example
# Create some dummy correlated data
data <- AutoQuant::FakeDataGenerator(
Correlation = 0.85,
N = 10000,
ID = 2,
ZIP = 0,
AddDate = FALSE,
Classification = FALSE,
MultiClass = FALSE)
# Copy data
data1 <- data.table::copy(data)
# Define features names
Features <- c(names(data1)[!names(data1) %in% c('IDcol_1','IDcol_2','Adrian')])
# Run function
ModelObject <- AutoQuant::AutoCatBoostRegression(
# GPU or CPU and the number of available GPUs
task_type = 'GPU',
NumGPUs = 1,
NumOfParDepPlots = length(Features),
# Metadata args
OutputSelection = c('Importances','EvalPlots','EvalMetrics','Score_TrainData'),
ModelID = 'Test_Model_1',
model_path = getwd(),
metadata_path = getwd(),
ReturnModelObjects = TRUE,
# Data args
data = data1,
TargetColumnName = 'Adrian',
FeatureColNames = Features,
IDcols = c('IDcol_1','IDcol_2'),
TransformNumericColumns = 'Adrian',
Methods = c('Asinh','Asin','Log','LogPlus1','Sqrt','Logit'))
# Build report
AutoQuant::ModelInsightsReport(
# Meta info
TargetColumnName = 'Adrian',
PredictionColumnName = 'Predict',
FeatureColumnNames = Features,
DateColumnName = NULL,
# Control options
TargetType = 'regression',
ModelID = 'Test_Model_1',
Algo = 'catboost',
OutputPath = getwd(),
ModelObject = ModelObject)Classification ModelInsightsReport() Example
# Create some dummy correlated data
data <- AutoQuant::FakeDataGenerator(
Correlation = 0.85,
N = 10000,
ID = 2,
ZIP = 0,
AddDate = FALSE,
Classification = TRUE,
MultiClass = FALSE)
# Copy data
data1 <- data.table::copy(data)
# Feature names
Features <- c(names(data1)[!names(data1) %in% c('IDcol_1','IDcol_2','Adrian')])
# Run function
ModelObject <- AutoQuant::AutoCatBoostClassifier(
# GPU or CPU and the number of available GPUs
task_type = 'GPU',
NumGPUs = 1,
# Metadata args
OutputSelection = c('Score_TrainData', 'Importances', 'EvalPlots', 'EvalMetrics'),
ModelID = 'Test_Model_1',
model_path = getwd(),
metadata_path = getwd(),
ReturnModelObjects = TRUE,
NumOfParDepPlots = length(Features),
# Data args
data = data1,
TargetColumnName = 'Adrian',
FeatureColNames = Features,
IDcols = c('IDcol_1','IDcol_2'))
# Build report
AutoQuant::ModelInsightsReport(
# Meta info
TargetColumnName = 'Adrian',
PredictionColumnName = 'p1',
FeatureColumnNames = Features,
DateColumnName = NULL,
# Control options
TargetType = 'classification',
ModelID = 'Test_Model_1',
Algo = 'catboost',
OutputPath = getwd(),
ModelObject = ModelObject)MultiClass ModelInsightsReport() Example
# Create some dummy correlated data
data <- AutoQuant::FakeDataGenerator(
Correlation = 0.85,
N = 10000L,
ID = 2L,
ZIP = 0L,
AddDate = FALSE,
Classification = FALSE,
MultiClass = TRUE)
# Copy data
data1 <- data.table::copy(data)
# Feature Colnames
Features <- c(names(data1)[!names(data1) %in% c('IDcol_1','IDcol_2','Adrian')])
# Run function
ModelObject <- AutoQuant::AutoCatBoostMultiClass(
# GPU or CPU and the number of available GPUs
task_type = 'GPU',
NumGPUs = 1,
NumOfParDepPlots = length(Features),
# Metadata args
OutputSelection = c('Importances', 'EvalPlots', 'EvalMetrics', 'Score_TrainData'),
ModelID = 'Test_Model_1',
model_path = getwd(),
metadata_path = getwd(),
ReturnModelObjects = TRUE,
# Data args
data = data,
TargetColumnName = 'Adrian',
FeatureColNames = Features,
IDcols = c('IDcol_1','IDcol_2'))
# Create Model Insights Report
AutoQuant::ModelInsightsReport(
# DataSets (use TestData for ValidationData)
TrainData = train,
ValidationData = valid,
TestData = test,
# Meta info
TargetColumnName = 'Adrian',
PredictionColumnName = 'Predict',
FeatureColumnNames = Features,
# Control options
TargetType = 'MultiClass',
ModelID = 'Test_Model_1',
Algo = 'catboost',
OutputPath = getwd(),
ModelObject = ModelObject)Expand to view content
Code Example: AutoCatBoostCARMA()
# @@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@
# Out-of-Sample Feature + Grid Tuning of AutoQuant::AutoCatBoostCARMA()
# @@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@
# Set up your output file path for saving results as a .csv
Path <- "C:/YourPathHere"
# Run on GPU or CPU (some options in the grid tuning force usage of CPU for some runs)
TaskType = "GPU"
# Define number of CPU threads to allow data.table to utilize
data.table::setDTthreads(percent = max(1L, parallel::detectCores()-2L))
# Load data
data <- data <- data.table::fread("https://www.dropbox.com/s/2str3ek4f4cheqi/walmart_train.csv?dl=1")
# Ensure series have no missing dates (also remove series with more than 25% missing values)
data <- AutoQuant::TimeSeriesFill(
data,
DateColumnName = "Date",
GroupVariables = c("Store","Dept"),
TimeUnit = "weeks",
FillType = "maxmax",
MaxMissingPercent = 0.25,
SimpleImpute = TRUE)
# Set negative numbers to 0
data <- data[, Weekly_Sales := data.table::fifelse(Weekly_Sales < 0, 0, Weekly_Sales)]
# Remove IsHoliday column
data[, IsHoliday := NULL]
# Create xregs (this is the include the categorical variables instead of utilizing only the interaction of them)
xregs <- data[, .SD, .SDcols = c("Date", "Store", "Dept")]
# Change data types
data[, ":=" (Store = as.character(Store), Dept = as.character(Dept))]
xregs[, ":=" (Store = as.character(Store), Dept = as.character(Dept))]
# Subset data so we have an out of time sample
data1 <- data.table::copy(data[, ID := 1L:.N, by = c("Store","Dept")][ID <= 125L][, ID := NULL])
data[, ID := NULL]
# Define values for SplitRatios and FCWindow Args
N1 <- data1[, .N, by = c("Store","Dept")][1L, N]
N2 <- xregs[, .N, by = c("Store","Dept")][1L, N]
# Setup Grid Tuning & Feature Tuning data.table using a cross join of vectors
Tuning <- data.table::CJ(
TimeWeights = c("None",0.999),
MaxTimeGroups = c("weeks","months"),
TargetTransformation = c("TRUE","FALSE"),
Difference = c("TRUE","FALSE"),
HoldoutTrain = c(6,18),
Langevin = c("TRUE","FALSE"),
NTrees = c(2500,5000),
Depth = c(6,9),
RandomStrength = c(0.75,1),
L2_Leaf_Reg = c(3.0,4.0),
RSM = c(0.75,"NULL"),
GrowPolicy = c("SymmetricTree","Lossguide","Depthwise"),
BootStrapType = c("Bayesian","MVS","No"))
# Remove options that are not compatible with GPU (skip over this otherwise)
Tuning <- Tuning[Langevin == "TRUE" | (Langevin == "FALSE" & RSM == "NULL" & BootStrapType %in% c("Bayesian","No"))]
# Randomize order of Tuning data.table
Tuning <- Tuning[order(runif(.N))]
# Load grid results and remove rows that have already been tested
if(file.exists(file.path(Path, "Walmart_CARMA_Metrics.csv"))) {
Metrics <- data.table::fread(file.path(Path, "Walmart_CARMA_Metrics.csv"))
temp <- data.table::rbindlist(list(Metrics,Tuning), fill = TRUE)
temp <- unique(temp, by = c(4:(ncol(temp)-1)))
Tuning <- temp[is.na(RunTime)][, .SD, .SDcols = names(Tuning)]
rm(Metrics,temp)
}
# Define the total number of runs
TotalRuns <- Tuning[,.N]
# Kick off feature + grid tuning
for(Run in seq_len(TotalRuns)) {
# Print run number
for(zz in seq_len(100)) print(Run)
# Use fresh data for each run
xregs_new <- data.table::copy(xregs)
data_new <- data.table::copy(data1)
# Timer start
StartTime <- Sys.time()
# Run carma system
CatBoostResults <- AutoQuant::AutoCatBoostCARMA(
# data args
data = data_new,
TimeWeights = if(Tuning[Run, TimeWeights] == "None") NULL else as.numeric(Tuning[Run, TimeWeights]),
TargetColumnName = "Weekly_Sales",
DateColumnName = "Date",
HierarchGroups = NULL,
GroupVariables = c("Store","Dept"),
TimeUnit = "weeks",
TimeGroups = if(Tuning[Run, MaxTimeGroups] == "weeks") "weeks" else if(Tuning[Run, MaxTimeGroups] == "months") c("weeks","months") else c("weeks","months","quarters"),
# Production args
TrainOnFull = TRUE,
SplitRatios = c(1 - Tuning[Run, HoldoutTrain] / N2, Tuning[Run, HoldoutTrain] / N2),
PartitionType = "random",
FC_Periods = N2-N1,
TaskType = TaskType,
NumGPU = 1,
Timer = TRUE,
DebugMode = TRUE,
# Target variable transformations
TargetTransformation = as.logical(Tuning[Run, TargetTransformation]),
Methods = c('Asinh','Log','LogPlus1','Sqrt'),
Difference = as.logical(Tuning[Run, Difference]),
NonNegativePred = TRUE,
RoundPreds = FALSE,
# Calendar-related features
CalendarVariables = c("week","wom","month","quarter"),
HolidayVariable = c("USPublicHolidays"),
HolidayLookback = NULL,
HolidayLags = c(1,2,3),
HolidayMovingAverages = c(2,3),
# Lags, moving averages, and other rolling stats
Lags = if(Tuning[Run, MaxTimeGroups] == "weeks") c(1,2,3,4,5,8,9,12,13,51,52,53) else if(Tuning[Run, MaxTimeGroups] == "months") list("weeks" = c(1,2,3,4,5,8,9,12,13,51,52,53), "months" = c(1,2,6,12)) else list("weeks" = c(1,2,3,4,5,8,9,12,13,51,52,53), "months" = c(1,2,6,12), "quarters" = c(1,2,3,4)),
MA_Periods = if(Tuning[Run, MaxTimeGroups] == "weeks") c(2,3,4,5,8,9,12,13,51,52,53) else if(Tuning[Run, MaxTimeGroups] == "months") list("weeks" = c(2,3,4,5,8,9,12,13,51,52,53), "months" = c(2,6,12)) else list("weeks" = c(2,3,4,5,8,9,12,13,51,52,53), "months" = c(2,6,12), "quarters" = c(2,3,4)),
SD_Periods = NULL,
Skew_Periods = NULL,
Kurt_Periods = NULL,
Quantile_Periods = NULL,
Quantiles_Selected = NULL,
# Bonus features
AnomalyDetection = NULL,
XREGS = xregs_new,
FourierTerms = 0,
TimeTrendVariable = TRUE,
ZeroPadSeries = NULL,
DataTruncate = FALSE,
# ML grid tuning args
GridTune = FALSE,
PassInGrid = NULL,
ModelCount = 5,
MaxRunsWithoutNewWinner = 50,
MaxRunMinutes = 60*60,
# ML evaluation output
PDFOutputPath = NULL,
SaveDataPath = NULL,
NumOfParDepPlots = 0L,
# ML loss functions
EvalMetric = "RMSE",
EvalMetricValue = 1,
LossFunction = "RMSE",
LossFunctionValue = 1,
# ML tuning args
NTrees = Tuning[Run, NTrees],
Depth = Tuning[Run, Depth],
L2_Leaf_Reg = Tuning[Run, L2_Leaf_Reg],
LearningRate = 0.03,
Langevin = as.logical(Tuning[Run, Langevin]),
DiffusionTemperature = 10000,
RandomStrength = Tuning[Run, RandomStrength],
BorderCount = 254,
RSM = if(Tuning[Run, RSM] == "NULL") NULL else as.numeric(Tuning[Run, RSM]),
GrowPolicy = Tuning[Run, GrowPolicy],
BootStrapType = Tuning[Run, BootStrapType],
ModelSizeReg = 0.5,
FeatureBorderType = "GreedyLogSum",
SamplingUnit = "Group",
SubSample = NULL,
ScoreFunction = "Cosine",
MinDataInLeaf = 1)
# Timer End
EndTime <- Sys.time()
# Prepare data for evaluation
Results <- CatBoostResults$Forecast
data.table::setnames(Results, "Weekly_Sales", "bla")
Results <- merge(Results, data, by = c("Store","Dept","Date"), all = FALSE)
Results <- Results[is.na(bla)][, bla := NULL]
# Create totals and subtotals
Results <- data.table::groupingsets(
x = Results,
j = list(Predictions = sum(Predictions), Weekly_Sales = sum(Weekly_Sales)),
by = c("Date", "Store", "Dept"),
sets = list(c("Date", "Store", "Dept"), c("Store", "Dept"), "Store", "Dept", "Date"))
# Fill NAs with "Total" for totals and subtotals
for(cols in c("Store","Dept")) Results[, eval(cols) := data.table::fifelse(is.na(get(cols)), "Total", get(cols))]
# Add error measures
Results[, Weekly_MAE := abs(Weekly_Sales - Predictions)]
Results[, Weekly_MAPE := Weekly_MAE / Weekly_Sales]
# Weekly results
Weekly_MAPE <- Results[, list(Weekly_MAPE = mean(Weekly_MAPE)), by = list(Store,Dept)]
# Monthly results
temp <- data.table::copy(Results)
temp <- temp[, Date := lubridate::floor_date(Date, unit = "months")]
temp <- temp[, lapply(.SD, sum), by = c("Date","Store","Dept"), .SDcols = c("Predictions", "Weekly_Sales")]
temp[, Monthly_MAE := abs(Weekly_Sales - Predictions)]
temp[, Monthly_MAPE := Monthly_MAE / Weekly_Sales]
Monthly_MAPE <- temp[, list(Monthly_MAPE = mean(Monthly_MAPE)), by = list(Store,Dept)]
# Collect metrics for Total (feel free to switch to something else or no filter at all)
Metrics <- data.table::data.table(
RunNumber = Run,
Total_Weekly_MAPE = Weekly_MAPE[Store == "Total" & Dept == "Total", Weekly_MAPE],
Total_Monthly_MAPE = Monthly_MAPE[Store == "Total" & Dept == "Total", Monthly_MAPE],
Tuning[Run],
RunTime = EndTime - StartTime)
# Append to file (not overwrite)
data.table::fwrite(Metrics, file = file.path(Path, "Walmart_CARMA_Metrics.csv"), append = TRUE)
# Remove objects (clear space before new runs)
rm(CatBoostResults, Results, temp, Weekly_MAE, Weekly_MAPE, Monthly_MAE, Monthly_MAPE)
# Garbage collection because of GPU
gc()
}Code Example: AutoXGBoostCARMA()
# @@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@
# XGBoost Version ----
# @@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@
# Load data
data <- data.table::fread("https://www.dropbox.com/s/2str3ek4f4cheqi/walmart_train.csv?dl=1")
# Ensure series have no missing dates (also remove series with more than 25% missing values)
data <- AutoQuant::TimeSeriesFill(
data,
DateColumnName = "Date",
GroupVariables = c("Store","Dept"),
TimeUnit = "weeks",
FillType = "maxmax",
MaxMissingPercent = 0.25,
SimpleImpute = TRUE)
# Set negative numbers to 0
data <- data[, Weekly_Sales := data.table::fifelse(Weekly_Sales < 0, 0, Weekly_Sales)]
# Remove IsHoliday column
data[, IsHoliday := NULL]
# Create xregs (this is the include the categorical variables instead of utilizing only the interaction of them)
xregs <- data[, .SD, .SDcols = c("Date", "Store", "Dept")]
# Change data types
data[, ":=" (Store = as.character(Store), Dept = as.character(Dept))]
xregs[, ":=" (Store = as.character(Store), Dept = as.character(Dept))]
# Build forecast
XGBoostResults <- AutoXGBoostCARMA(
# Data Artifacts
data = data,
NonNegativePred = FALSE,
RoundPreds = FALSE,
TargetColumnName = "Weekly_Sales",
DateColumnName = "Date",
HierarchGroups = NULL,
GroupVariables = c("Store","Dept"),
TimeUnit = "weeks",
TimeGroups = c("weeks","months"),
# Data Wrangling Features
ZeroPadSeries = NULL,
DataTruncate = FALSE,
SplitRatios = c(1 - 10 / 138, 10 / 138),
PartitionType = "timeseries",
AnomalyDetection = NULL,
EncodingMethod = "binary",
# Productionize
FC_Periods = 0,
TrainOnFull = FALSE,
NThreads = 8,
Timer = TRUE,
DebugMode = FALSE,
SaveDataPath = NULL,
PDFOutputPath = NULL,
# Target Transformations
TargetTransformation = TRUE,
Methods = c("Asinh", "Asin", "Log", "LogPlus1", "Sqrt", "Logit"),
Difference = FALSE,
# Features
Lags = list("weeks" = seq(1L, 10L, 1L), "months" = seq(1L, 5L, 1L)),
MA_Periods = list("weeks" = seq(5L, 20L, 5L), "months" = seq(2L, 10L, 2L)),
SD_Periods = NULL,
Skew_Periods = NULL,
Kurt_Periods = NULL,
Quantile_Periods = NULL,
Quantiles_Selected = c("q5","q95"),
XREGS = xregs,
FourierTerms = 4,
CalendarVariables = c("week", "wom", "month", "quarter"),
HolidayVariable = c("USPublicHolidays","EasterGroup","ChristmasGroup","OtherEcclesticalFeasts"),
HolidayLookback = NULL,
HolidayLags = 1,
HolidayMovingAverages = 1:2,
TimeTrendVariable = TRUE,
# ML eval args
TreeMethod = "hist",
EvalMetric = "RMSE",
LossFunction = 'reg:squarederror',
# ML grid tuning
GridTune = FALSE,
ModelCount = 5,
MaxRunsWithoutNewWinner = 20L,
MaxRunMinutes = 24L*60L,
# ML args
NTrees = 300,
LearningRate = 0.3,
MaxDepth = 9L,
MinChildWeight = 1.0,
SubSample = 1.0,
ColSampleByTree = 1.0)Code Example: AutoLightGBMCARMA()
# @@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@
# LightGBM Version ----
# @@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@
# Load data
data <- data.table::fread('https://www.dropbox.com/s/2str3ek4f4cheqi/walmart_train.csv?dl=1')
# Ensure series have no missing dates (also remove series with more than 25% missing values)
data <- AutoQuant::TimeSeriesFill(
data,
DateColumnName = 'Date',
GroupVariables = c('Store','Dept'),
TimeUnit = 'weeks',
FillType = 'maxmax',
MaxMissingPercent = 0.25,
SimpleImpute = TRUE)
# Set negative numbers to 0
data <- data[, Weekly_Sales := data.table::fifelse(Weekly_Sales < 0, 0, Weekly_Sales)]
# Remove IsHoliday column
data[, IsHoliday := NULL]
# Create xregs (this is the include the categorical variables instead of utilizing only the interaction of them)
xregs <- data[, .SD, .SDcols = c('Date', 'Store', 'Dept')]
# Change data types
data[, ':=' (Store = as.character(Store), Dept = as.character(Dept))]
xregs[, ':=' (Store = as.character(Store), Dept = as.character(Dept))]
# Build forecast
Results <- AutoLightGBMCARMA(
# Data Artifacts
data = data,
NonNegativePred = FALSE,
RoundPreds = FALSE,
TargetColumnName = 'Weekly_Sales',
DateColumnName = 'Date',
HierarchGroups = NULL,
GroupVariables = c('Store','Dept'),
TimeUnit = 'weeks',
TimeGroups = c('weeks','months'),
# Data Wrangling Features
EncodingMethod = 'binary',
ZeroPadSeries = NULL,
DataTruncate = FALSE,
SplitRatios = c(1 - 10 / 138, 10 / 138),
PartitionType = 'timeseries',
AnomalyDetection = NULL,
# Productionize
FC_Periods = 0,
TrainOnFull = FALSE,
NThreads = 8,
Timer = TRUE,
DebugMode = FALSE,
SaveDataPath = NULL,
PDFOutputPath = NULL,
# Target Transformations
TargetTransformation = TRUE,
Methods = c('Asinh', 'Asin', 'Log', 'LogPlus1', 'Sqrt', 'Logit'),
Difference = FALSE,
# Features
Lags = list('weeks' = seq(1L, 10L, 1L), 'months' = seq(1L, 5L, 1L)),
MA_Periods = list('weeks' = seq(5L, 20L, 5L), 'months' = seq(2L, 10L, 2L)),
SD_Periods = NULL,
Skew_Periods = NULL,
Kurt_Periods = NULL,
Quantile_Periods = NULL,
Quantiles_Selected = c('q5','q95'),
XREGS = xregs,
FourierTerms = 4,
CalendarVariables = c('week', 'wom', 'month', 'quarter'),
HolidayVariable = c('USPublicHolidays','EasterGroup','ChristmasGroup','OtherEcclesticalFeasts'),
HolidayLookback = NULL,
HolidayLags = 1,
HolidayMovingAverages = 1:2,
TimeTrendVariable = TRUE,
# ML eval args
TreeMethod = 'hist',
EvalMetric = 'RMSE',
LossFunction = 'reg:squarederror',
# Grid tuning args
GridTune = FALSE,
GridEvalMetric = 'mae',
ModelCount = 30L,
MaxRunsWithoutNewWinner = 20L,
MaxRunMinutes = 24L*60L,
# LightGBM Args
Device_Type = TaskType,
LossFunction = 'regression',
EvalMetric = 'MAE',
Input_Model = NULL,
Task = 'train',
Boosting = 'gbdt',
LinearTree = FALSE,
Trees = 1000,
ETA = 0.10,
Num_Leaves = 31,
Deterministic = TRUE,
# Learning Parameters
# https://lightgbm.readthedocs.io/en/latest/Parameters.html#learning-control-parameters
Force_Col_Wise = FALSE,
Force_Row_Wise = FALSE,
Max_Depth = 6,
Min_Data_In_Leaf = 20,
Min_Sum_Hessian_In_Leaf = 0.001,
Bagging_Freq = 1.0,
Bagging_Fraction = 1.0,
Feature_Fraction = 1.0,
Feature_Fraction_Bynode = 1.0,
Lambda_L1 = 0.0,
Lambda_L2 = 0.0,
Extra_Trees = FALSE,
Early_Stopping_Round = 10,
First_Metric_Only = TRUE,
Max_Delta_Step = 0.0,
Linear_Lambda = 0.0,
Min_Gain_To_Split = 0,
Drop_Rate_Dart = 0.10,
Max_Drop_Dart = 50,
Skip_Drop_Dart = 0.50,
Uniform_Drop_Dart = FALSE,
Top_Rate_Goss = FALSE,
Other_Rate_Goss = FALSE,
Monotone_Constraints = NULL,
Monotone_Constraints_Method = 'advanced',
Monotone_Penalty = 0.0,
Forcedsplits_Filename = NULL, # use for AutoStack option; .json file
Refit_Decay_Rate = 0.90,
Path_Smooth = 0.0,
# IO Dataset Parameters
# https://lightgbm.readthedocs.io/en/latest/Parameters.html#io-parameters
Max_Bin = 255,
Min_Data_In_Bin = 3,
Data_Random_Seed = 1,
Is_Enable_Sparse = TRUE,
Enable_Bundle = TRUE,
Use_Missing = TRUE,
Zero_As_Missing = FALSE,
Two_Round = FALSE,
# Convert Parameters
Convert_Model = NULL,
Convert_Model_Language = 'cpp',
# Objective Parameters
# https://lightgbm.readthedocs.io/en/latest/Parameters.html#objective-parameters
Boost_From_Average = TRUE,
Alpha = 0.90,
Fair_C = 1.0,
Poisson_Max_Delta_Step = 0.70,
Tweedie_Variance_Power = 1.5,
Lambdarank_Truncation_Level = 30,
# Metric Parameters (metric is in Core)
# https://lightgbm.readthedocs.io/en/latest/Parameters.html#metric-parameters
Is_Provide_Training_Metric = TRUE,
Eval_At = c(1,2,3,4,5),
# Network Parameters
# https://lightgbm.readthedocs.io/en/latest/Parameters.html#network-parameters
Num_Machines = 1,
# GPU Parameters
# https://lightgbm.readthedocs.io/en/latest/Parameters.html#gpu-parameters
Gpu_Platform_Id = -1,
Gpu_Device_Id = -1,
Gpu_Use_Dp = TRUE,
Num_Gpu = 1)Code Example: AutoH2OCARMA()
# @@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@
# H2O Version ----
# @@@@@@@@@@@@@@@@@@@@@@@@@@@@@@@
# Load data
data <- data.table::fread("https://www.dropbox.com/s/2str3ek4f4cheqi/walmart_train.csv?dl=1")
# Ensure series have no missing dates (also remove series with more than 25% missing values)
data <- AutoQuant::TimeSeriesFill(
data,
DateColumnName = "Date",
GroupVariables = c("Store","Dept"),
TimeUnit = "weeks",
FillType = "maxmax",
MaxMissingPercent = 0.25,
SimpleImpute = TRUE)
# Set negative numbers to 0
data <- data[, Weekly_Sales := data.table::fifelse(Weekly_Sales < 0, 0, Weekly_Sales)]
# Remove IsHoliday column
data[, IsHoliday := NULL]
# Create xregs (this is the include the categorical variables instead of utilizing only the interaction of them)
xregs <- data[, .SD, .SDcols = c("Date", "Store", "Dept")]
# Change data types
data[, ":=" (Store = as.character(Store), Dept = as.character(Dept))]
xregs[, ":=" (Store = as.character(Store), Dept = as.character(Dept))]
# Build forecast
Results <- AutoQuant::AutoH2OCARMA(
# Data Artifacts
AlgoType = "drf",
ExcludeAlgos = NULL,
data = data,
TargetColumnName = "Weekly_Sales",
DateColumnName = "Date",
HierarchGroups = NULL,
GroupVariables = c("Dept"),
TimeUnit = "week",
TimeGroups = c("weeks","months"),
# Data Wrangling Features
SplitRatios = c(1 - 10 / 138, 10 / 138),
PartitionType = "random",
# Production args
FC_Periods = 4L,
TrainOnFull = FALSE,
MaxMem = {gc();paste0(as.character(floor(max(32, as.numeric(system("awk '/MemFree/ {print $2}' /proc/meminfo", intern=TRUE)) -32) / 1000000)),"G")},
NThreads = parallel::detectCores(),
PDFOutputPath = NULL,
SaveDataPath = NULL,
Timer = TRUE,
DebugMode = TRUE,
# Target Transformations
TargetTransformation = FALSE,
Methods = c("Asinh", "Asin", "Log", "LogPlus1", "Sqrt", "Logit"),
Difference = FALSE,
NonNegativePred = FALSE,
RoundPreds = FALSE,
# Calendar features
CalendarVariables = c("week", "wom", "month", "quarter", "year"),
HolidayVariable = c("USPublicHolidays","EasterGroup",
"ChristmasGroup","OtherEcclesticalFeasts"),
HolidayLookback = NULL,
HolidayLags = 1:7,
HolidayMovingAverages = 2:7,
TimeTrendVariable = TRUE,
# Time series features
Lags = list("weeks" = c(1:4), "months" = c(1:3)),
MA_Periods = list("weeks" = c(2:8), "months" = c(6:12)),
SD_Periods = NULL,
Skew_Periods = NULL,
Kurt_Periods = NULL,
Quantile_Periods = NULL,
Quantiles_Selected = NULL,
# Bonus Features
XREGS = NULL,
FourierTerms = 2L,
AnomalyDetection = NULL,
ZeroPadSeries = NULL,
DataTruncate = FALSE,
# ML evaluation args
EvalMetric = "RMSE",
NumOfParDepPlots = 0L,
# ML grid tuning args
GridTune = FALSE,
GridStrategy = "Cartesian",
ModelCount = 5,
MaxRuntimeSecs = 60*60*24,
StoppingRounds = 10,
# ML Args
NTrees = 1000L,
MaxDepth = 20,
SampleRate = 0.632,
MTries = -1,
ColSampleRatePerTree = 1,
ColSampleRatePerTreeLevel = 1,
MinRows = 1,
NBins = 20,
NBinsCats = 1024,
NBinsTopLevel = 1024,
HistogramType = "AUTO",
CategoricalEncoding = "AUTO",
RandomColNumbers = NULL,
InteractionColNumbers = NULL,
WeightsColumn = NULL,
# ML args
Distribution = "gaussian",
Link = "identity",
RandomDistribution = NULL,
RandomLink = NULL,
Solver = "AUTO",
Alpha = NULL,
Lambda = NULL,
LambdaSearch = FALSE,
NLambdas = -1,
Standardize = TRUE,
RemoveCollinearColumns = FALSE,
InterceptInclude = TRUE,
NonNegativeCoefficients = FALSE)
Expand to view content
Time Series Forecasting Description
There are three sets of functions for single series traditional time series model forecasting. The first set includes the AutoBanditSarima() and AutoBanditNNet() functions. These two offer the most robust fitting strategies. The utilize a multi-armed-bandit to help narrow the search space of available parameter settings. The next batch includes the AutoTBATS(), AutoETS(), and the AutoArfima() functions. These don't utilze the bandit framework. Rather, they run through a near exhaustive search through all their possible settings. Both the bandit set and the non-bandit set utilize parallelism to burn through as many models as possible for a fixed amount of time.
- Bandit: AutoBanditSarima
- Bandit: AutoBanditNNet
- Exhaustive: AutoTBATS
- Exhaustive: AutoETS
- Exhaustive: AutoArfima
AutoBanditSarima() Example
# Build model
data <- AutoQuant::FakeDataGenerator(Correlation = 0.82, TimeSeries = TRUE, TimeSeriesTimeAgg = "1min")
# Run system
Output <- AutoQuant::AutoBanditSarima(
data = data,
SaveFile = NULL,
ByDataType = FALSE,
TargetVariableName = "Weekly_Sales",
DateColumnName = "Date",
TimeAggLevel = "1min",
EvaluationMetric = "MAE",
NumHoldOutPeriods = 12L,
NumFCPeriods = 16L,
MaxLags = 10L,
MaxSeasonalLags = 0L,
MaxMovingAverages = 3L,
MaxSeasonalMovingAverages = 0L,
MaxFourierPairs = 2L,
TrainWeighting = 0.50,
MaxConsecutiveFails = 50L,
MaxNumberModels = 100L,
MaxRunTimeMinutes = 10L,
NumberCores = 12,
DebugMode = FALSE)
# View output
Output$ForecastPlot
Output$ErrorLagMA2x2
Output$Forecast
Output$PerformanceGridAutoBanditNNet() Example
# Build model
data <- AutoQuant::FakeDataGenerator(Correlation = 0.82, TimeSeries = TRUE, TimeSeriesTimeAgg = "1min")
# Run system
Output <- AutoQuant::AutoBanditNNet(
data = data,
TargetVariableName = "Weekly_Sales",
DateColumnName = "Date",
TimeAggLevel = "1min",
EvaluationMetric = "MAE",
NumHoldOutPeriods = 12L,
NumFCPeriods = 16L,
MaxLags = 10L,
MaxSeasonalLags = 0L,
MaxFourierPairs = 2L,
TrainWeighting = 0.50,
MaxConsecutiveFails = 50L,
MaxNumberModels = 100L,
MaxRunTimeMinutes = 10L,
NumberCores = 12)
# View output
Output$Forecast
Output$PerformanceGridAutoTBATS() Example
# Build model
data <- AutoQuant::FakeDataGenerator(Correlation = 0.82, TimeSeries = TRUE, TimeSeriesTimeAgg = "1min")
# Run system
Output <- AutoQuant::AutoTBATS(
data = data,
FilePath = getwd(),
TargetVariableName = "Weekly_Sales",
DateColumnName = "Date",
TimeAggLevel = "1min",
EvaluationMetric = "MAE",
NumHoldOutPeriods = 12L,
NumFCPeriods = 16L,
MaxLags = 10L,
MaxMovingAverages = 5,
MaxSeasonalPeriods = 1,
TrainWeighting = 0.50,
MaxConsecutiveFails = 50L,
MaxNumberModels = 100L,
MaxRunTimeMinutes = 10L,
NumberCores = 12)
# View output
Output$Forecast
Output$PerformanceGridAutoETS() Example
# Build model
data <- AutoQuant::FakeDataGenerator(Correlation = 0.82, TimeSeries = TRUE, TimeSeriesTimeAgg = "1min")
# Run system
Output <- AutoQuant::AutoETS(
data = data,
FilePath = getwd(),
TargetVariableName = "Weekly_Sales",
DateColumnName = "Date",
TimeAggLevel = "1min",
EvaluationMetric = "MAE",
NumHoldOutPeriods = 12L,
NumFCPeriods = 16L,
TrainWeighting = 0.50,
MaxConsecutiveFails = 50L,
MaxNumberModels = 100L,
MaxRunTimeMinutes = 10L,
NumberCores = 12)
# View output
Output$Forecast
Output$PerformanceGridAutoArfima() Example
# Build model
data <- AutoQuant::FakeDataGenerator(Correlation = 0.82, TimeSeries = TRUE, TimeSeriesTimeAgg = "1min")
# Run system
Output <- AutoQuant::AutoArfima(
data = data,
FilePath = getwd(),
TargetVariableName = "Weekly_Sales",
DateColumnName = "Date",
TimeAggLevel = "1min",
EvaluationMetric = "MAE",
NumHoldOutPeriods = 12L,
NumFCPeriods = 16L,
TrainWeighting = 0.50,
MaxLags = 5,
MaxMovingAverages = 5,
MaxConsecutiveFails = 50L,
MaxNumberModels = 100L,
MaxRunTimeMinutes = 10L,
NumberCores = 12)
# View output
Output$Forecast
Output$PerformanceGrid