Comments (26)
For the text, you can pick an x and then ax.text(x, y, "37.3") but might have to turn off clipping at boundaries so it appears. Or, set xlim to 20% more and turn off tick marks/labels for that part then ax.text() there. beat it into submission!! haha
I had to get the wedges below the x axis and had to turn off clipping to do that. you can check the wedge code in my stuff for that.
from dtreeviz.
For spacing, i think just shrink total height of graph maybe?
It shrinks a little, but there is still a lot of space. Because the standard deviation between dots is sigma=0.08, with mean = 0.5 (mu value). leaf index are from 1 to 1... tomorrow, I will play with different values for them.
from dtreeviz.
wow! now that is a damn fine looking plot, sir! I will merge!
from dtreeviz.
I'm not sure if i understood correctly. Would you like to include all the leaf split plots into a single new visualisation ?
Something similar with this ? :)

from dtreeviz.
yep, that's it!!! Seems like it'd be useful just as the stacked histo is for classification
from dtreeviz.
Cool ! Do you have any web resources/experience of how to include subplots (leaf sample'y distribution) into another plot using matplotlib ?
from dtreeviz.
hahaha. i spent forever learning how to do that in this project. we shouldn't have to though. We can create a single plot that just does scattering with some noise for each leaf area. Here is sample code:
mu = .5
sigma = .08
X = np.random.normal(mu, sigma, size=len(y))
ax.set_xlim(0, 1)
alpha = colors['scatter_marker_alpha'] # was .25
ax.scatter(X, y, s=5, c=colors['scatter_marker'], alpha=alpha, lw=.3)
ax.plot([0,len(node.samples())],[m,m],'--', color=colors['split_line'], linewidth=1)
So, basically, go to each leaf id and do a scatter that is x = id+noise and the y is just the y of samples at that leaf. make sense?
from dtreeviz.
Nicee, I got the idea. I will start to implement it in the next days.
from dtreeviz.
While thinking about this new visualisation, I have remember the boxplot from matplotlib. I guess it's what we need : https://matplotlib.org/3.1.1/gallery/statistics/boxplot_demo.html.
It can show a lot of build in stats, like median, mean, quartiles, outliers, etc (being a common visualisation in matplotlib, could be easier to interpret by other people)
@parrt what do you think ?
from dtreeviz.
Only problem is that box plots are ugly ;) The alpha channel helps density and human eye is really good at picking up spread of data.
from dtreeviz.
Indeed, box plot are a little ugly :)). I will take a look at the code that you mentioned
from dtreeviz.
I have an intermediate visualisation.
Here is how it looks for a tree with depth=3:

depth=5:

depth=10:

As we can see, the visualisations become hard to read/interpret when we have a lot of leaves.
@parrt do you have any feedback, suggestions ?
from dtreeviz.
looking interesting. for one we should tighten and fix the x direction so it's always the same. also dots should be bigger and then alpha channel can make darker areas show higher density. maybe graph is a bit too tall too. :) Maybe we add a show arg like rtree_univar that turns off x labels? Or maybe just show prediction to make smaller?
from dtreeviz.
- 'for one we should tighten and fix the x direction so it's always the same.'
Do you mean to hardcode a fix value for figsize argument ? - 'also dots should be bigger and then alpha channel can make darker areas show higher density'
agree, we can also make the markersize configurable - 'Maybe we add a show arg like rtree_univar that turns off x labels? '
agree - Or maybe just show prediction to make smaller?
Do you mean to show only the horizontal black lines ?
from dtreeviz.
@parrt could be an option to show the leaves in multiple plots when the number of leaves is big ?
from dtreeviz.
Looking good! Yes, i suppose we could split up but that's harder to implement. maybe args that say leaf ID range min/max?
- 'for one we should tighten and fix the x direction so it's always the same.'
Do you mean to hardcode a fix value for figsize argument ?
I just meant that any specific leaf split plot should be same width; don't smear it out to fit graph width. Graph width is function of how many leaves I think. - 'also dots should be bigger and then alpha channel can make darker areas show higher density'
agree, we can also make the markersize configurable
yep - 'Maybe we add a show arg like rtree_univar that turns off x labels? '
agree
cool - Or maybe just show prediction to make smaller?
Do you mean to show only the horizontal black lines ?
That would work. I was earlier thinking that the x labels beneath graph would show just prediction value rather than num samples as well. just to make smaller.
from dtreeviz.
I created a PR. We can add modifications on it.
I added the option to create more plots, with a specified number of leaves/plot (plot_leaf_count)
For 1 :
- I added a configurable figsize for the plot. In case of multiple plots, the last plot will keep the same width for leaves as others plots.
- Done
- in the end, I added show_leaf_labels bool parameter (similar with dtreeviz method), it seems to me more straightforward than show parameter (in case I don't want to display the x labels, I would have to initialize show=[]).
- removed the samples from the string.
from dtreeviz.
Cool. What about just 20.4 not age=20.4 to make even smaller. Then make age the overall x label?
from dtreeviz.
actually that might look weird like it's the x-value when in fact it's the leaf id. hmm...what if we put the predicted value at the top and then we don't actually need an x tick label? We'd just need "leaf id" or something for the grpah xlabel and then "age" for ylabel.
from dtreeviz.
I created the horizontal implementation for visualisation. I like how it looks, it's a better option than multiple plots. I also added a configurable 'grid' parameter, which shows the grid in visualisation (it helps to identify leaf's dots).
I feel that we need to came up with better values for x, y labels (like you pointed up in previous comment).
from dtreeviz.
Yeah, certainly repeating the target name is not optimal. I'd suggest something like:
Also let's try to keep things tightened up with perhaps less vertical space between leaves?
from dtreeviz.
"Yeah, certainly repeating the target name is not optimal. I'd suggest something like:"
I will make a little research how to put the label in the right side of y axis.
"Also let's try to keep things tightened up with perhaps less vertical space between leaves?"
For this I will need to adjust the 'mu' and 'sigma' values and maybe the leaf index...
from dtreeviz.
For spacing, i think just shrink total height of graph maybe?
from dtreeviz.
- reduced the space between leaves. Created a parameter for these also (leaf_space)
- about y axis, I put "leaf prediction" as a label and leaf predictions for y ticks. I looks well for me, it makes the graph loaded with few information. @parrt what do you think ? :)
from dtreeviz.
Looking better! I think it's confusing to see prediction on y. that should be "leaf" I think as you had it. :) also maybe double thick the prediction bars? Perhaps your split plots have a too-high variance, maybe tighten up more like
The idea is that it's kind of a "shaky line of dots". Might have to reduce alpha channel a bit depending on number of dots.
from dtreeviz.
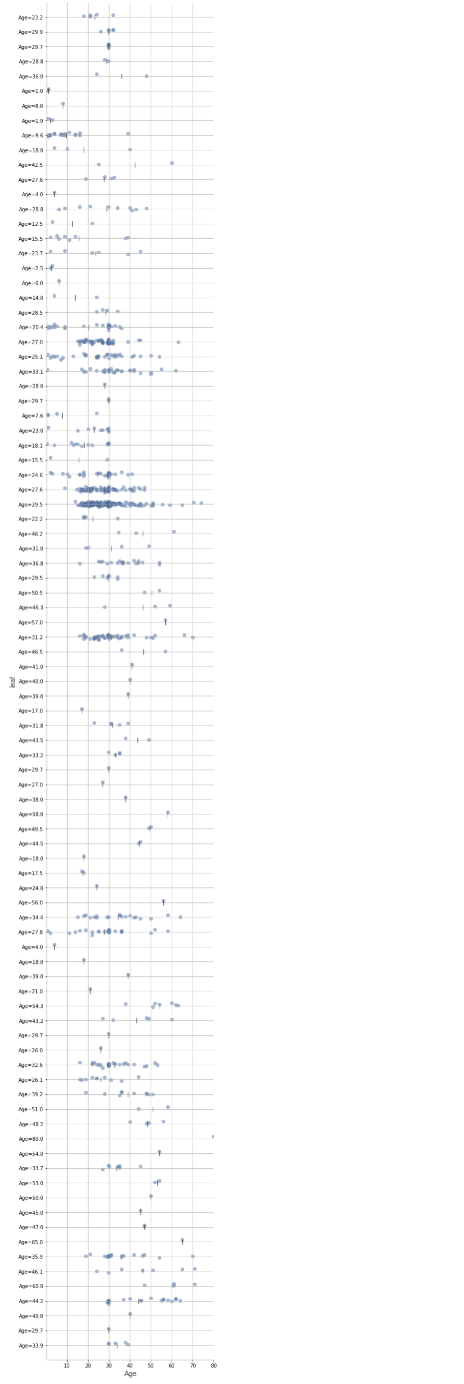
hi @parrt.
- I added the y label in the right side of the plot.
- narrow the leaf dots visualisations
- double the prediction bars
Here it is how they look now :)
from dtreeviz.
Related Issues (20)
- from dtreeviz.trees import * Import necessary libraries HOT 1
- Color keyword argument - Value error HOT 14
- Add support for TensorFlow GradientBoostedTreesModel model
- _regr_leaf_viz calculates the mean for prediction value.
- WARNING:matplotlib.font_manager:findfont: Font family 'Arial' not found. HOT 2
- Decision Tree visualize wrong path HOT 1
- When using dataset that is different from the training for trees models - does not draw HOT 1
- Support for RandomForest HOT 5
- Visualize custom decision tree HOT 1
- how to use dtreeviz in streamlit HOT 2
- VisualisationNotYetSupportedError: get_min_samples_leaf() is not implemented yet for XGBoost. HOT 4
- TypeError: list indices must be integers or slices, not numpy.float64 HOT 5
- Crash when leaf nodes have no samples HOT 1
- Out of memory when calling viz.view() HOT 2
- Integrate AI explanation
- CatBoost need to be supported. HOT 1
- AttributeError: module 'dtreeviz' has no attribute 'model' on Windows platform, works fine on Google colab
- tfdf.keras.CartModel support? HOT 1
- TypeError: 'int' object is not subscriptable HOT 3
- Development requirement in `setup.py` HOT 1
Recommend Projects
-
 React
React
A declarative, efficient, and flexible JavaScript library for building user interfaces.
-
Vue.js
🖖 Vue.js is a progressive, incrementally-adoptable JavaScript framework for building UI on the web.
-
 Typescript
Typescript
TypeScript is a superset of JavaScript that compiles to clean JavaScript output.
-
TensorFlow
An Open Source Machine Learning Framework for Everyone
-
Django
The Web framework for perfectionists with deadlines.
-
Laravel
A PHP framework for web artisans
-
D3
Bring data to life with SVG, Canvas and HTML. 📊📈🎉
-
Recommend Topics
-
javascript
JavaScript (JS) is a lightweight interpreted programming language with first-class functions.
-
web
Some thing interesting about web. New door for the world.
-
server
A server is a program made to process requests and deliver data to clients.
-
Machine learning
Machine learning is a way of modeling and interpreting data that allows a piece of software to respond intelligently.
-
Visualization
Some thing interesting about visualization, use data art
-
Game
Some thing interesting about game, make everyone happy.
Recommend Org
-
Facebook
We are working to build community through open source technology. NB: members must have two-factor auth.
-
Microsoft
Open source projects and samples from Microsoft.
-
Google
Google ❤️ Open Source for everyone.
-
Alibaba
Alibaba Open Source for everyone
-
D3
Data-Driven Documents codes.
-
Tencent
China tencent open source team.











from dtreeviz.